Dissecting the role of MPS1 in chromosome biorientation and the spindle checkpoint through the small molecule inhibitor reversine
- PMID: 20624901
- PMCID: PMC2911657
- DOI: 10.1083/jcb.201001036
Dissecting the role of MPS1 in chromosome biorientation and the spindle checkpoint through the small molecule inhibitor reversine
Abstract
The catalytic activity of the MPS1 kinase is crucial for the spindle assembly checkpoint and for chromosome biorientation on the mitotic spindle. We report that the small molecule reversine is a potent mitotic inhibitor of MPS1. Reversine inhibits the spindle assembly checkpoint in a dose-dependent manner. Its addition to mitotic HeLa cells causes the ejection of Mad1 and the ROD-ZWILCH-ZW10 complex, both of which are important for the spindle checkpoint, from unattached kinetochores. By using reversine, we also demonstrate that MPS1 is required for the correction of improper chromosome-microtubule attachments. We provide evidence that MPS1 acts downstream from the AURORA B kinase, another crucial component of the error correction pathway. Our experiments describe a very useful tool to interfere with MPS1 activity in human cells. They also shed light on the relationship between the error correction pathway and the spindle checkpoint and suggest that these processes are coregulated and are likely to share at least a subset of their catalytic machinery.
Figures
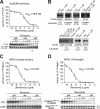


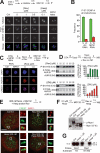


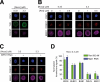

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

