Surprising gene expression patterns within and between PDF-containing circadian neurons in Drosophila
- PMID: 20624977
- PMCID: PMC2922133
- DOI: 10.1073/pnas.1002081107
Surprising gene expression patterns within and between PDF-containing circadian neurons in Drosophila
Abstract
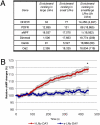
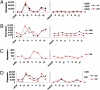
To compare circadian gene expression within highly discrete neuronal populations, we separately purified and characterized two adjacent but distinct groups of Drosophila adult circadian neurons: the 8 small and 10 large PDF-expressing ventral lateral neurons (s-LNvs and l-LNvs, respectively). The s-LNvs are the principal circadian pacemaker cells, whereas recent evidence indicates that the l-LNvs are involved in sleep and light-mediated arousal. Although half of the l-LNv-enriched mRNA population, including core clock mRNAs, is shared between the l-LNvs and s-LNvs, the other half is l-LNv- and s-LNv-specific. The distribution of four specific mRNAs is consistent with prior characterization of the four encoded proteins, and therefore indicates successful purification of the two neuronal types. Moreover, an octopamine receptor mRNA is selectively enriched in l-LNvs, and only these neurons respond to in vitro application of octopamine. Dissection and purification of l-LNvs from flies collected at different times indicate that these neurons contain cycling clock mRNAs with higher circadian amplitudes as well as at least a 10-fold higher fraction of oscillating mRNAs than all previous analyses of head RNA. Many of these cycling l-LNv mRNAs are well expressed but do not cycle or cycle much less well elsewhere in heads. The results suggest that RNA cycling is much more prominent in circadian neurons than elsewhere in heads and may be particularly important for the functioning of these neurons.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Hardin PE. Transcription regulation within the circadian clock: The E-box and beyond. J Biol Rhythms. 2004;19:348–360. - PubMed
-
- Rosbash M, et al. Transcriptional feedback and definition of the circadian pacemaker in Drosophila and animals. Cold Spring Harb Symp Quant Biol. 2007;72:75–83. - PubMed
-
- Price JL, et al. double-time is a novel Drosophila clock gene that regulates PERIOD protein accumulation. Cell. 1998;94:83–95. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

