CDK5RAP2 regulates centriole engagement and cohesion in mice
- PMID: 20627074
- PMCID: PMC3078807
- DOI: 10.1016/j.devcel.2010.05.017
CDK5RAP2 regulates centriole engagement and cohesion in mice
Abstract
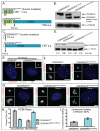
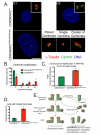
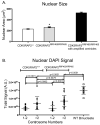
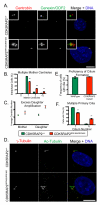
Centriole duplication occurs once per cell cycle, ensuring that each cell contains two centrosomes, each containing a mother-daughter pair of tightly engaged centrioles at mitotic entry. Loss of the tight engagement between mother and daughter centrioles appears to license the next round of centriole duplication. However, the molecular mechanisms regulating this process remain largely unknown. Mutations in CDK5RAP2, which encodes a centrosomal protein, cause autosomal recessive primary microcephaly in humans. Here we show that CDK5RAP2 loss of function in mice causes centriole amplification with a preponderance of single, unpaired centrioles and increased numbers of daughter-daughter centriole pairs. These results indicate that CDK5RAP2 is required to maintain centriole engagement and cohesion, thereby restricting centriole replication. Early in mitosis, amplified centrosomes assemble multipolar spindles in CDK5RAP2 mutant cells. Moreover, both mother and daughter centrioles are amplified and the excess mother centrioles template multiple primary cilia in CDK5RAP2 mutant cells.
Copyright 2010 Elsevier Inc. All rights reserved.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

