A genome-wide siRNA screen reveals multiple mTORC1 independent signaling pathways regulating autophagy under normal nutritional conditions
- PMID: 20627085
- PMCID: PMC2935848
- DOI: 10.1016/j.devcel.2010.05.005
A genome-wide siRNA screen reveals multiple mTORC1 independent signaling pathways regulating autophagy under normal nutritional conditions
Abstract
Autophagy is a cellular catabolic mechanism that plays an essential function in protecting multicellular eukaryotes from neurodegeneration, cancer, and other diseases. However, we still know very little about mechanisms regulating autophagy under normal homeostatic conditions when nutrients are not limiting. In a genome-wide human siRNA screen, we demonstrate that under normal nutrient conditions upregulation of autophagy requires the type III PI3 kinase, but not inhibition of mTORC1, the essential negative regulator of starvation-induced autophagy. We show that a group of growth factors and cytokines inhibit the type III PI3 kinase through multiple pathways, including the MAPK-ERK1/2, Stat3, Akt/Foxo3, and CXCR4/GPCR, which are all known to positively regulate cell growth and proliferation. Our study suggests that the type III PI3 kinase integrates diverse signals to regulate cellular levels of autophagy, and that autophagy and cell proliferation may represent two alternative cell fates that are regulated in a mutually exclusive manner.
Copyright 2010 Elsevier Inc. All rights reserved.
Figures

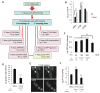
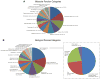


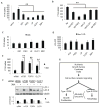
Comment in
-
The autophagy connection.Dev Cell. 2010 Jul 20;19(1):11-2. doi: 10.1016/j.devcel.2010.07.005. Dev Cell. 2010. PMID: 20643346 Free PMC article.
References
-
- Byfield MP, Murray JT, Backer JM. hVps34 is a nutrient-regulated lipid kinase required for activation of p70 S6 kinase. J Biol Chem. 2005;280:33076–33082. - PubMed
-
- Chung N, Zhang XD, Kreamer A, Locco L, Kuan PF, Bartz S, Linsley PS, Ferrer M, Strulovici B. Median absolute deviation to improve hit selection for genome-scale RNAi screens. J Biomol Screen. 2008;13:149–158. - PubMed
-
- Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, Yokoyama M, Mishima K, Saito I, Okano H, et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–889. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

