MicroRNA 320a functions as a novel endogenous modulator of aquaporins 1 and 4 as well as a potential therapeutic target in cerebral ischemia
- PMID: 20628061
- PMCID: PMC2937953
- DOI: 10.1074/jbc.M110.144576
MicroRNA 320a functions as a novel endogenous modulator of aquaporins 1 and 4 as well as a potential therapeutic target in cerebral ischemia
Abstract
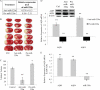
Aquaporins facilitate efficient diffusion of water across cellular membranes, and water homeostasis is critically important in conditions such as cerebral edema. Changes in aquaporin 1 and 4 expression in the brain are associated with cerebral edema, and the lack of water channel modulators is often highlighted. Here we present evidence of an endogenous modulator of aquaporin 1 and 4. We identify miR-320a as a potential modulator of aquaporin 1 and 4 and explore the possibility of using miR-320a to alter the expression of aquaporin 1 and 4 in normal and ischemic conditions. We show that precursor miR-320a can function as an inhibitor, whereas anti-miR-320a can act as an activator of aquaporin 1 and 4 expressions. We have also shown that anti-miR-320a could bring about a reduction of infarct volume in cerebral ischemia with a concomitant increase in aquaporins 1 and 4 mRNA and protein expression.
Figures

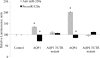
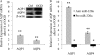
Similar articles
-
MicroRNA-130a represses transcriptional activity of aquaporin 4 M1 promoter.J Biol Chem. 2012 Apr 6;287(15):12006-15. doi: 10.1074/jbc.M111.280701. Epub 2012 Feb 13. J Biol Chem. 2012. PMID: 22334710 Free PMC article.
-
miR-320a affects spinal cord edema through negatively regulating aquaporin-1 of blood-spinal cord barrier during bimodal stage after ischemia reperfusion injury in rats.BMC Neurosci. 2016 Feb 5;17:10. doi: 10.1186/s12868-016-0243-1. BMC Neurosci. 2016. Retraction in: BMC Neurosci. 2021 Nov 3;22(1):64. doi: 10.1186/s12868-021-00669-6. PMID: 26850728 Free PMC article. Retracted.
-
Aquaporin 4 changes in rat brain with severe hydrocephalus.Eur J Neurosci. 2006 Jun;23(11):2929-36. doi: 10.1111/j.1460-9568.2006.04829.x. Eur J Neurosci. 2006. PMID: 16819982
-
Role of aquaporins in cell migration and edema formation in human brain tumors.Exp Cell Res. 2011 Oct 15;317(17):2391-6. doi: 10.1016/j.yexcr.2011.07.006. Epub 2011 Jul 20. Exp Cell Res. 2011. PMID: 21784068 Review.
-
Aquaporins in Cardiovascular System.Adv Exp Med Biol. 2017;969:105-113. doi: 10.1007/978-94-024-1057-0_6. Adv Exp Med Biol. 2017. PMID: 28258568 Review.
Cited by
-
Cell biology of ischemia/reperfusion injury.Int Rev Cell Mol Biol. 2012;298:229-317. doi: 10.1016/B978-0-12-394309-5.00006-7. Int Rev Cell Mol Biol. 2012. PMID: 22878108 Free PMC article. Review.
-
Aquaporins and Ion Channels as Dual Targets in the Design of Novel Glioblastoma Therapeutics to Limit Invasiveness.Cancers (Basel). 2023 Jan 30;15(3):849. doi: 10.3390/cancers15030849. Cancers (Basel). 2023. PMID: 36765806 Free PMC article. Review.
-
MicroRNAs in the Blood-Brain Barrier in Hypoxic-Ischemic Brain Injury.Curr Neuropharmacol. 2020;18(12):1180-1186. doi: 10.2174/1570159X18666200429004242. Curr Neuropharmacol. 2020. PMID: 32348227 Free PMC article. Review.
-
The role of miR-320a and IL-1β in human chondrocyte degradation.Bone Joint Res. 2017 Apr;6(4):196-203. doi: 10.1302/2046-3758.64.BJR-2016-0224.R1. Bone Joint Res. 2017. PMID: 28404547 Free PMC article.
-
Differential expression of non-coding RNAs and association with cerebral ischemic vascular disorders; diagnostic and therapeutic opportunities.Genes Genomics. 2023 Jul;45(7):835-845. doi: 10.1007/s13258-022-01281-6. Epub 2022 Jul 8. Genes Genomics. 2023. PMID: 35802344 Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

