Partial loss of ataxin-1 function contributes to transcriptional dysregulation in spinocerebellar ataxia type 1 pathogenesis
- PMID: 20628574
- PMCID: PMC2900305
- DOI: 10.1371/journal.pgen.1001021
Partial loss of ataxin-1 function contributes to transcriptional dysregulation in spinocerebellar ataxia type 1 pathogenesis
Abstract
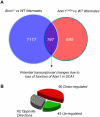
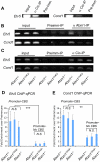
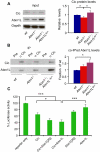
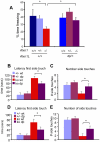
Spinocerebellar ataxia type 1 (SCA1) is a dominantly inherited neurodegenerative disease caused by expansion of a CAG repeat that encodes a polyglutamine tract in ATAXIN1 (ATXN1). Molecular and genetic data indicate that SCA1 is mainly caused by a gain-of-function mechanism. However, deletion of wild-type ATXN1 enhances SCA1 pathogenesis, whereas increased levels of an evolutionarily conserved paralog of ATXN1, Ataxin 1-Like, ameliorate it. These data suggest that a partial loss of ATXN1 function contributes to SCA1. To address this possibility, we set out to determine if the SCA1 disease model (Atxn1(154Q/+) mice) and the loss of Atxn1 function model (Atxn1-/- mice) share molecular changes that could potentially contribute to SCA1 pathogenesis. To identify transcriptional changes that might result from loss of function of ATXN1 in SCA1, we performed gene expression microarray studies on cerebellar RNA from Atxn1-/- and Atxn1(154Q/+) cerebella and uncovered shared gene expression changes. We further show that mild overexpression of Ataxin-1-Like rescues several of the molecular and behavioral defects in Atxn1-/- mice. These results support a model in which Ataxin 1-Like overexpression represses SCA1 pathogenesis by compensating for a partial loss of function of Atxn1. Altogether, these data provide evidence that partial loss of Atxn1 function contributes to SCA1 pathogenesis and raise the possibility that loss-of-function mechanisms contribute to other dominantly inherited neurodegenerative diseases.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
-
- Orr HT, Zoghbi HY. Trinucleotide repeat disorders. Annu Rev Neurosci. 2007;30:575–621. - PubMed
-
- Gatchel JR, Zoghbi HY. Diseases of unstable repeat expansion: mechanisms and common principles. Nat Rev Genet. 2005;6:743–755. - PubMed
-
- Zuhlke C, Riess O, Bockel B, Lange H, Thies U. Mitotic stability and meiotic variability of the (CAG)n repeat in the Huntington disease gene. Hum Mol Genet. 1993;2:2063–2067. - PubMed
-
- Keckarevic D, Culjkovic B, Savic D, Stojkovic O, Kostic V, et al. The status of SCA1, MJD/SCA3, FRDA, DRPLA and MD triplet containing genes in patients with Huntington disease and healthy controls. J Neurogenet. 2000;14:257–263. - PubMed
-
- Lorenzetti D, Bohlega S, Zoghbi HY. The expansion of the CAG repeat in ataxin-2 is a frequent cause of autosomal dominant spinocerebellar ataxia. Neurology. 1997;49:1009–1013. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- NS22920/NS/NINDS NIH HHS/United States
- NS45667/NS/NINDS NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- R01 NS022920/NS/NINDS NIH HHS/United States
- P30 HD024064/HD/NICHD NIH HHS/United States
- R37 NS022920/NS/NINDS NIH HHS/United States
- NS27699/NS/NINDS NIH HHS/United States
- T32 NS043124/NS/NINDS NIH HHS/United States
- R01 NS045667/NS/NINDS NIH HHS/United States
- R37 NS027699/NS/NINDS NIH HHS/United States
- P30HD024064/HD/NICHD NIH HHS/United States
- F31 NS052925/NS/NINDS NIH HHS/United States
- R01 NS027699/NS/NINDS NIH HHS/United States
- 1F31NS052925/NS/NINDS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

