Dynamics of HIV-1 quasispecies during antiviral treatment dissected using ultra-deep pyrosequencing
- PMID: 20628644
- PMCID: PMC2898805
- DOI: 10.1371/journal.pone.0011345
Dynamics of HIV-1 quasispecies during antiviral treatment dissected using ultra-deep pyrosequencing
Abstract
Background: Ultra-deep pyrosequencing (UDPS) allows identification of rare HIV-1 variants and minority drug resistance mutations, which are not detectable by standard sequencing.
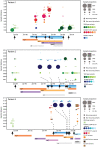
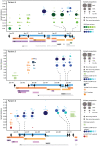
Principal findings: Here, UDPS was used to analyze the dynamics of HIV-1 genetic variation in reverse transcriptase (RT) (amino acids 180-220) in six individuals consecutively sampled before, during and after failing 3TC and AZT containing antiretroviral treatment. Optimized UDPS protocols and bioinformatic software were developed to generate, clean and analyze the data. The data cleaning strategy reduced the error rate of UDPS to an average of 0.05%, which is lower than previously reported. Consequently, the cut-off for detection of resistance mutations was very low. A median of 16,016 (range 2,406-35,401) sequence reads were obtained per sample, which allowed detection and quantification of minority resistance mutations at amino acid position 181, 184, 188, 190, 210, 215 and 219 in RT. In four of five pre-treatment samples low levels (0.07-0.09%) of the M184I mutation were observed. Other resistance mutations, except T215A and T215I were below the detection limit. During treatment failure, M184V replaced M184I and dominated the population in combination with T215Y, while wild-type variants were rarely detected. Resistant virus disappeared rapidly after treatment interruption and was undetectable as early as after 3 months. In most patients, drug resistant variants were replaced by wild-type variants identical to those present before treatment, suggesting rebound from latent reservoirs.
Conclusions: With this highly sensitive UDPS protocol preexisting drug resistance was infrequently observed; only M184I, T215A and T215I were detected at very low levels. Similarly, drug resistant variants in plasma quickly decreased to undetectable levels after treatment interruption. The study gives important insights into the dynamics of the HIV-1 quasispecies and is of relevance for future research and clinical use of the UDPS technology.
Conflict of interest statement
Figures

References
-
- Eigen M, Schuster P. The hypercycle. A principle of natural self-organization. Part A: Emergence of the hypercycle. Naturwissenschaften. 1977;64:541–565. - PubMed
-
- Meyerhans A, Cheynier R, Albert J, Seth M, Kwok S, et al. Temporal fluctuations in HIV quasispecies in vivo are not reflected by sequential HIV isolations. Cell. 1989;58:901–910. - PubMed
-
- Preston BD. Reverse transcriptase fidelity and HIV-1 variation. Science. 1997;275:228–229; author reply 230-221. - PubMed
-
- Coffin JM. HIV population dynamics in vivo: implications for genetic variation, pathogenesis, and therapy. Science. 1995;267:483–489. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

