Novel insights into proteolytic cleavage of influenza virus hemagglutinin
- PMID: 20629046
- PMCID: PMC7169116
- DOI: 10.1002/rmv.657
Novel insights into proteolytic cleavage of influenza virus hemagglutinin
Abstract
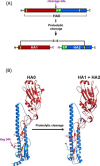
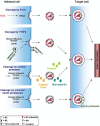
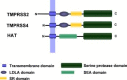
The influenza virus hemagglutinin (HA) mediates the first essential step in the viral life cycle, virus entry into target cells. Influenza virus HA is synthesised as a precursor protein in infected cells and requires cleavage by host cell proteases to transit into an active form. Cleavage is essential for influenza virus infectivity and the HA-processing proteases are attractive targets for therapeutic intervention. It is well established that cleavage by ubiquitously expressed subtilisin-like proteases is a hallmark of highly pathogenic avian influenza viruses (HPAIV). In contrast, the nature of the proteases responsible for cleavage of HA of human influenza viruses and low pathogenic avian influenza viruses (LPAIV) is not well understood. Recent studies suggest that cleavage of HA of human influenza viruses might be a cell-associated event and might be facilitated by the type II transmembrane serine proteases (TTSPs) TMPRSS2, TMPRSS4 and human airway trypsin-like protease (HAT). Here, we will introduce the different concepts established for proteolytic activation of influenza virus HA, with a particular focus on the role of TTSPs, and we will discuss their implications for viral tropism, pathogenicity and antiviral intervention.
(c) 2010 John Wiley & Sons, Ltd.
Figures
References
-
- Palese P. Influenza: old and new threats. Nat Med 2004; 10: S82–S87. - PubMed
-
- Thompson WW, Shay DK, Weintraub E, et al. Mortality associated with influenza and respiratory syncytial virus in the United States. J Am Med Assoc 2003; 289: 179–186. - PubMed
-
- Belshe RB. The origins of pandemic influenza–lessons from the 1918 virus. N Engl J Med 2005; 353: 2209–2211. - PubMed
-
- Parrish CR, Kawaoka Y. The origins of new pandemic viruses: the acquisition of new host ranges by canine parvovirus and influenza A viruses. Annu Rev Microbiol 2005; 59: 553–586. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

