Identification of primary gene targets of TFAP2C in hormone responsive breast carcinoma cells
- PMID: 20629094
- PMCID: PMC2928401
- DOI: 10.1002/gcc.20807
Identification of primary gene targets of TFAP2C in hormone responsive breast carcinoma cells
Abstract
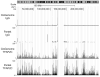
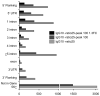
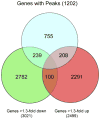
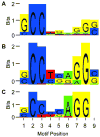
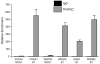
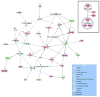
The TFAP2C transcription factor is involved in mammary development, differentiation, and oncogenesis. Previous studies established a role for TFAP2C in the regulation of ESR1 (ERalpha) and ERBB2 (Her2) in breast carcinomas. However, the role of TFAP2C in different breast cancer phenotypes has not been examined in detail. To develop a more complete characterization of TFAP2C target genes, ChIP-seq with anti-TFAP2C antibody and expression arrays with TFAP2C knock down were analyzed in MCF-7 breast carcinoma cells. Genomic sequences common to the ChIP-seq data set defined the consensus sequence for TFAP2C chromatin binding as the nine base sequence SCCTSRGGS (S = G/C, r = A/G), which closely matches the previously defined optimal in vitro binding site. Comparing expression arrays before and after knock down of TFAP2C with ChIP-seq data demonstrated a conservative estimate that 8% of genes altered by TFAP2C expression are primary target genes and includes genes that are both induced and repressed by TFAP2C. A set of 447 primary target genes of TFAP2C was identified, which included ESR1 (ERalpha), FREM2, RET, FOXA1, WWOX, GREB1, MYC, and members of the retinoic acid response pathway. The identification of ESR1, WWOX, GREB1, and FOXA1 as primary targets confirmed the role of TFAP2C in hormone response. TFAP2C plays a critical role in gene regulation in hormone responsive breast cancer and its target genes are different than for the Her2 breast cancer phenotype.
Figures


References
-
- Allouche A, Nolens G, Tancredi A, Delacroix L, Mardaga J, Fridman V, Winkler R, Boniver J, Delvenne P, Begon DY. The combined immunodetection of AP-2alpha and YY1 transcription factors is associated with ERBB2 gene overexpression in primary breast tumors. Breast Cancer Res. 2008;10:R9. - PMC - PubMed
-
- Begon DY, Delacroix L, Vernimmen D, Jackers P, Winkler R. Yin Yang 1 cooperates with activator protein 2 to stimulate ERBB2 gene expression in mammary cancer cells. J Biol Chem. 2005;280:24428–34. - PubMed
-
- Bosher JM, Totty NF, Hsuan JJ, Williams T, Hurst HC. A family of AP-2 proteins regulates c-erbB-2 expression in mammary carcinoma. Oncogene. 1996;13:1701–7. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

