Complete genome sequence of a Megalocytivirus (family Iridoviridae) associated with turbot mortality in China
- PMID: 20630106
- PMCID: PMC2912838
- DOI: 10.1186/1743-422X-7-159
Complete genome sequence of a Megalocytivirus (family Iridoviridae) associated with turbot mortality in China
Abstract
Background: Turbot reddish body iridovirus (TRBIV) causes serious systemic diseases with high mortality in the cultured turbot, Scophthalmus maximus. We here sequenced and analyzed the complete genome of TRBIV, which was identified in Shandong province, China.
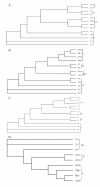
Results: The genome of TRBIV is a linear double-stranded DNA of 110,104 base pairs, comprising 55% G + C. Total 115 open reading frames were identified, encoding polypeptides ranging from 40 to 1168 amino acids. Amino acid sequences analysis revealed that 39 of the 115 potential gene products of TRBIV show significant homology to other iridovirus proteins. Phylogenetic analysis of conserved genes indicated that TRBIV is closely related to infectious spleen and kidney necrosis virus (ISKNV), rock bream iridovirus (RBIV), orange-spotted grouper iridovirus (OSGIV), and large yellow croaker iridovirus (LYCIV). The results indicated that TRBIV belongs to the genus Megalocytivirus (family Iridoviridae).
Conclusions: The determination of the genome of TRBIV will provide useful information for comparative study of Megalocytivirus and developing strategies to control outbreaks of TRBIV-induced disease.
Figures

References
-
- Williams T. The iridoviruses. Adv Virus Res. 1996;46:345–412. full_text. - PubMed
Publication types
MeSH terms
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

