Changes in colon gene expression associated with increased colon inflammation in interleukin-10 gene-deficient mice inoculated with Enterococcus species
- PMID: 20630110
- PMCID: PMC2912833
- DOI: 10.1186/1471-2172-11-39
Changes in colon gene expression associated with increased colon inflammation in interleukin-10 gene-deficient mice inoculated with Enterococcus species
Abstract
Background: Inappropriate responses to normal intestinal bacteria may be involved in the development of Inflammatory Bowel Diseases (IBD, e.g. Crohn's Disease (CD), Ulcerative Colitis (UC)) and variations in the host genome may mediate this process. IL-10 gene-deficient (Il10-/-) mice develop CD-like colitis mainly in the colon, in part due to inappropriate responses to normal intestinal bacteria including Enterococcus strains, and have therefore been used as an animal model of CD. Comprehensive characterization of changes in cecum gene expression levels associated with inflammation in the Il10-/- mouse model has recently been reported. Our aim was to characterize changes in colonic gene expression levels in Il10-/- and C57BL/6J (C57; control) mice resulting from oral bacterial inoculation with 12 Enterococcus faecalis and faecium (EF) strains isolated from calves or poultry, complex intestinal flora (CIF) collected from healthy control mice, or a mixture of the two (EF.CIF). We investigated two hypotheses: (1) that oral inoculation of Il10-/- mice would result in greater and more consistent intestinal inflammation than that observed in Il10-/- mice not receiving this inoculation, and (2) that this inflammation would be associated with changes in colon gene expression levels similar to those previously observed in human studies, and these mice would therefore be an appropriate model for human CD.
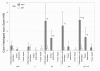
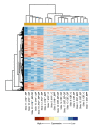
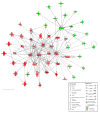
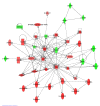
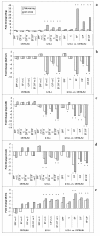
Results: At 12 weeks of age, total RNA extracted from intact colon was hybridized to Agilent 44 k mouse arrays. Differentially expressed genes were identified using linear models for microarray analysis (Bioconductor), and these genes were clustered using GeneSpring GX and Ingenuity Pathways Analysis software. Intestinal inflammation was increased in Il10-/- mice as a result of inoculation, with the strongest effect being in the EF and EF.CIF groups. Genes differentially expressed in Il10-/- mice as a result of EF or EF.CIF inoculation were associated with the following pathways: inflammatory disease (111 genes differentially expressed), immune response (209 genes), antigen presentation (11 genes, particularly major histocompatability complex Class II), fatty acid metabolism (30 genes) and detoxification (31 genes).
Conclusions: Our results suggest that colonic inflammation in Il10-/- mice inoculated with solutions containing Enterococcus strains is associated with gene expression changes similar to those of human IBD, specifically CD, and that with the EF.CIF inoculum in particular this is an appropriate model to investigate food-gene interactions relevant to human CD.
Figures

Similar articles
-
Nutrigenomics applied to an animal model of Inflammatory Bowel Diseases: transcriptomic analysis of the effects of eicosapentaenoic acid- and arachidonic acid-enriched diets.Mutat Res. 2007 Sep 1;622(1-2):103-16. doi: 10.1016/j.mrfmmm.2007.04.003. Epub 2007 Apr 19. Mutat Res. 2007. PMID: 17574631
-
Changes in composition of caecal microbiota associated with increased colon inflammation in interleukin-10 gene-deficient mice inoculated with Enterococcus species.Nutrients. 2015 Mar 11;7(3):1798-816. doi: 10.3390/nu7031798. Nutrients. 2015. PMID: 25768951 Free PMC article.
-
Inoculation with enterococci does not affect colon inflammation in the multi-drug resistance 1a-deficient mouse model of IBD.BMC Gastroenterol. 2016 Mar 3;16:31. doi: 10.1186/s12876-016-0447-y. BMC Gastroenterol. 2016. PMID: 26940566 Free PMC article.
-
Development, validation and implementation of an in vitro model for the study of metabolic and immune function in normal and inflamed human colonic epithelium.Dan Med J. 2015 Jan;62(1):B4973. Dan Med J. 2015. PMID: 25557335 Review.
-
The clinical, molecular, and therapeutic features of patients with IL10/IL10R deficiency: a systematic review.Clin Exp Immunol. 2022 Jun 23;208(3):281-291. doi: 10.1093/cei/uxac040. Clin Exp Immunol. 2022. PMID: 35481870 Free PMC article.
Cited by
-
Surface-Associated Lipoproteins Link Enterococcus faecalis Virulence to Colitogenic Activity in IL-10-Deficient Mice Independent of Their Expression Levels.PLoS Pathog. 2015 Jun 12;11(6):e1004911. doi: 10.1371/journal.ppat.1004911. eCollection 2015 Jun. PLoS Pathog. 2015. PMID: 26067254 Free PMC article.
-
Post-weaning selenium and folate supplementation affects gene and protein expression and global DNA methylation in mice fed high-fat diets.BMC Med Genomics. 2013 Mar 5;6:7. doi: 10.1186/1755-8794-6-7. BMC Med Genomics. 2013. PMID: 23497688 Free PMC article.
-
Role and mechanism of gut microbiota-host interactions in the pathogenesis of Crohn's disease.Int J Colorectal Dis. 2025 May 28;40(1):130. doi: 10.1007/s00384-025-04917-7. Int J Colorectal Dis. 2025. PMID: 40437310 Free PMC article. Review.
-
Complex Bacterial Consortia Reprogram the Colitogenic Activity of Enterococcus faecalis in a Gnotobiotic Mouse Model of Chronic, Immune-Mediated Colitis.Front Immunol. 2019 Jun 20;10:1420. doi: 10.3389/fimmu.2019.01420. eCollection 2019. Front Immunol. 2019. PMID: 31281321 Free PMC article.
-
A Multihit Model: Colitis Lessons from the Interleukin-10-deficient Mouse.Inflamm Bowel Dis. 2015 Aug;21(8):1967-75. doi: 10.1097/MIB.0000000000000468. Inflamm Bowel Dis. 2015. PMID: 26164667 Free PMC article. Review.
References
-
- de Buhr MF, Mahler M, Geffers R, Hansen W, Westendorf AM, Lauber J, Buer J, Schlegelberger B, Hedrich HJ, Bleich A. Cd14, Gbp1, and Pla2g2a: three major candidate genes for experimental IBD identified by combining QTL and microarray analyses. Physiol Genomics. 2006;25(3):426–434. doi: 10.1152/physiolgenomics.00022.2005. - DOI - PubMed
-
- Hampe J, Cuthbert A, Croucher PJ, Mirza MM, Mascheretti S, Fisher S, Frenzel H, King K, Hasselmeyer A, MacPherson AJ. Association between insertion mutation in NOD2 gene and Crohn's disease in German and British populations. Lancet. 2001;357(9272):1925–1928. doi: 10.1016/S0140-6736(00)05063-7. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

