A model in vitro system for co-transcriptional splicing
- PMID: 20631007
- PMCID: PMC2995048
- DOI: 10.1093/nar/gkq620
A model in vitro system for co-transcriptional splicing
Abstract
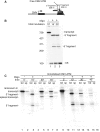
A hallmark of metazoan RNA polymerase II transcripts is the presence of numerous small exons surrounded by large introns. Abundant evidence indicates that splicing to excise introns occurs co-transcriptionally, prior to release of the nascent transcript from RNAP II. Here, we established an efficient model system for co-transcriptional splicing in vitro. In this system, CMV-DNA constructs immobilized on beads generate RNAP II transcripts containing two exons and an intron. Consistent with previous work, our data indicate that elongating nascent transcripts are tethered to RNAP II on the immobilized DNA template. We show that nascent transcripts that reach full length, but are still attached to RNAP II, are efficiently spliced. When the nascent transcript is cleaved within the intron using RNase H, both the 5' and 3' cleavage fragments are detected in the bound fraction, where they undergo splicing. Together, our work establishes a system for co-transcriptional splicing in vitro, in which the spliceosome containing the 5' and 3' exons are tethered to RNAP II for splicing.
Figures




References
-
- Black DL. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 2003;72:291–336. - PubMed

