Dopamine D1 receptors, regulation of gene expression in the brain, and neurodegeneration
- PMID: 20632973
- PMCID: PMC3803153
- DOI: 10.2174/187152710793361496
Dopamine D1 receptors, regulation of gene expression in the brain, and neurodegeneration
Abstract
Dopamine (DA), the most abundant catecholamine in the basal ganglia, participates in the regulation of motor functions and of cognitive processes such as learning and memory. Abnormalities in dopaminergic systems are thought to be the bases for some neuropsychiatric disorders including addiction, Parkinson's disease, and Schizophrenia. DA exerts its arrays of functions via stimulation of D1-like (D1 and D5) and D2-like (D2, D3, and D4) DA receptors which are located in various regions of the brain. The DA D1 and D2 receptors are very abundant in the basal ganglia where they exert their functions within separate neuronal cell types. The present paper focuses on a review of the effects of stimulation of DA D1 receptors on diverse signal transduction pathways and gene expression patterns in the brain. We also discuss the possible involvement of the DA D1 receptors in DA-mediated toxic effects observed both in vitro and in vivo. Future studies using more selective agonist and antagonist agents and the use of genetically modified animals should help to further clarify the role of these receptors in the normal physiology and in pathological events that involve DA.
Figures
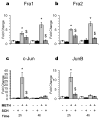
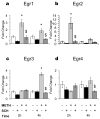
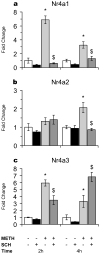
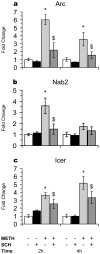
References
-
- Fallon JH. Topographic organization of ascending dopaminergic projections. Ann N Y Acad Sci. 1988;537:1–9. - PubMed
-
- Jimenez-Castellanos J, Graybiel AM. Subdivisions of the dopamine-containing A8-A9-A10 complex identified by their differential mesostriatal innervation of striosomes and extrastriosomal matrix. Neuroscience. 1987;23:223–242. - PubMed
-
- Bjorklund A, Dunnett SB. Dopamine neuron systems in the brain: an update. Trends Neurosci. 2007;30:194–202. - PubMed
-
- Chinta SJ, Andersen JK. Dopaminergic neurons. Int J Biochem Cell Biol. 2005;37:942–946. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
