Quantifying the mechanisms of domain gain in animal proteins
- PMID: 20633280
- PMCID: PMC2926785
- DOI: 10.1186/gb-2010-11-7-r74
Quantifying the mechanisms of domain gain in animal proteins
Abstract
Background: Protein domains are protein regions that are shared among different proteins and are frequently functionally and structurally independent from the rest of the protein. Novel domain combinations have a major role in evolutionary innovation. However, the relative contributions of the different molecular mechanisms that underlie domain gains in animals are still unknown. By using animal gene phylogenies we were able to identify a set of high confidence domain gain events and by looking at their coding DNA investigate the causative mechanisms.
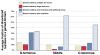
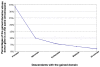
Results: Here we show that the major mechanism for gains of new domains in metazoan proteins is likely to be gene fusion through joining of exons from adjacent genes, possibly mediated by non-allelic homologous recombination. Retroposition and insertion of exons into ancestral introns through intronic recombination are, in contrast to previous expectations, only minor contributors to domain gains and have accounted for less than 1% and 10% of high confidence domain gain events, respectively. Additionally, exonization of previously non-coding regions appears to be an important mechanism for addition of disordered segments to proteins. We observe that gene duplication has preceded domain gain in at least 80% of the gain events.
Conclusions: The interplay of gene duplication and domain gain demonstrates an important mechanism for fast neofunctionalization of genes.
Figures



References
-
- Rubin GM, Yandell MD, Wortman JR, Gabor Miklos GL, Nelson CR, Hariharan IK, Fortini ME, Li PW, Apweiler R, Fleischmann W, Cherry JM, Henikoff S, Skupski MP, Misra S, Ashburner M, Birney E, Boguski MS, Brody T, Brokstein P, Celniker SE, Chervitz SA, Coates D, Cravchik A, Gabrielian A, Galle RF, Gelbart WM, George RA, Goldstein LS, Gong F, Guan P. Comparative genomics of the eukaryotes. Science. 2000;287:2204–2215. doi: 10.1126/science.287.5461.2204. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

