Transcriptional and translational effects of intronic CAPN3 gene mutations
- PMID: 20635405
- PMCID: PMC2966865
- DOI: 10.1002/humu.21320
Transcriptional and translational effects of intronic CAPN3 gene mutations
Abstract
Variants of unknown significance in the CAPN3 gene constitute a significant challenge for genetic counselling. Despite the frequency of intronic nucleotide changes in this gene (15-25% of all mutations), so far their pathogenicity has only been inferred by in-silico analysis, and occasionally, proven by RNA analysis. In this study, 5 different intronic variants (one novel) that bioinformatic tools predicted would affect RNA splicing, underwent comprehensive studies which were designed to prove they are disease-causing. Muscle mRNA from 15 calpainopathy patients was analyzed by RT-PCR and splicing-specific-PCR tests. We established the previously unrecognized pathogenicity of these mutations, which caused aberrant splicing, most frequently by the activation of cryptic splicing sites or, occasionally, by exon skipping. The absence or severe reduction of protein demonstrated their deleterious effect at translational level. We concluded that bioinformatic tools are valuable to suggest the potential effects of intronic variants; however, the experimental demonstration of the pathogenicity is not always easy to do even when using RNA analysis (low abundance, degradation mechanisms), and it might not be successful unless splicing-specific-PCR tests are used. A comprehensive approach is therefore recommended to identify and describe unclassified variants in order to offer essential data for basic and clinical geneticists.
Copyright 2010 Wiley-Liss, Inc.
Figures
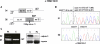
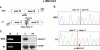


References
-
- Blázquez L, Azpitarte M, Sáenz A, Goicoechea M, Otaegui D, Ferrer X, Illa I, Gutierrez-Rivas E, Vilchez JJ, López de Munain A. Characterization of novel CAPN3 isoforms in white blood cells: an alternative approach for limb-girdle muscular dystrophy 2A diagnosis. Neurogenetics. 2008;9:173–182. - PubMed
-
- Duno M, Sveen ML, Schwartz M, Vissing J. cDNA analyses of CAPN3 enhance mutation detection and reveal a low prevalence of LGMD2A patients in Denmark. Eur J Hum Genet. 2008;16:935–940. - PubMed
-
- Fanin M, Nascimbeni AC, Aurino S, Tasca E, Pegoraro E, Nigro V, Angelini C. Frequency of LGMD gene mutations in Italian patients with distinct clinical phenotypes. Neurology. 2009a;72:1432–1435. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

