Mass spectrometry-based biomarker discovery: toward a global proteome index of individuality
- PMID: 20636062
- PMCID: PMC3140421
- DOI: 10.1146/annurev.anchem.1.031207.112942
Mass spectrometry-based biomarker discovery: toward a global proteome index of individuality
Abstract
Biomarker discovery and proteomics have become synonymous with mass spectrometry in recent years. Although this conflation is an injustice to the many essential biomolecular techniques widely used in biomarker-discovery platforms, it underscores the power and potential of contemporary mass spectrometry. Numerous novel and powerful technologies have been developed around mass spectrometry, proteomics, and biomarker discovery over the past 20 years to globally study complex proteomes (e.g., plasma). However, very few large-scale longitudinal studies have been carried out using these platforms to establish the analytical variability relative to true biological variability. The purpose of this review is not to cover exhaustively the applications of mass spectrometry to biomarker discovery, but rather to discuss the analytical methods and strategies that have been developed for mass spectrometry-based biomarker-discovery platforms and to place them in the context of the many challenges and opportunities yet to be addressed.
Figures
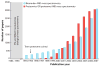

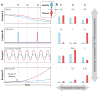

References
-
- Atkinson AJ, Colburn WA, DeGruttola VG, Demets DL, Downing GJ, et al. Biomarkers and surrogate endpoints: preferred definitions and conceptual framework. Clin Pharmacol Ther. 2001;69:89–95. - PubMed
-
- Elin RJ. Instrumentation in clinical chemistry. Science. 1980;210:286–89. - PubMed
-
- Chace DH. Mass spectrometry in the clinical laboratory. Chem Rev. 2001;101:445–77. - PubMed
-
- McPherson RA, Pincus MR. Henry’s Clinical Diagnosis and Management by Laboratory Methods. 21 Philadelphia: Saunders Elsevier; 2007. p. 1450.
-
- Burtis CA, Ashwood ER, editors. Tietz Fundamentals of Clinical Chemistry. 4 W.B. Saunders; 1996. p. 881.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

