Molecular analysis of the prokaryotic ubiquitin-like protein (Pup) conjugation pathway in Mycobacterium tuberculosis
- PMID: 20636328
- PMCID: PMC2975802
- DOI: 10.1111/j.1365-2958.2010.07276.x
Molecular analysis of the prokaryotic ubiquitin-like protein (Pup) conjugation pathway in Mycobacterium tuberculosis
Abstract
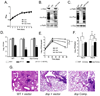
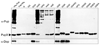
Proteins targeted for degradation by the Mycobacterium proteasome are post-translationally tagged with prokaryotic ubiquitin-like protein (Pup), an intrinsically disordered protein of 64 residues. In a process termed 'pupylation', Pup is synthesized with a terminal glutamine, which is deamidated to glutamate by Dop (deamidase of Pup) prior to attachment to substrate lysines by proteasome accessory factor A (PafA). Importantly, PafA was previously shown to be essential to cause lethal infections by Mycobacterium tuberculosis (Mtb) in mice. In this study we show that Dop, like PafA, is required for the full virulence of Mtb. Additionally, we show that Dop is not only involved in the deamidation of Pup, but also needed to maintain wild-type steady state levels of pupylated proteins in Mtb. Finally, using structural models and site-directed mutagenesis our data suggest that Dop and PafA are members of the glutamine synthetase fold family of proteins.
© 2010 Blackwell Publishing Ltd.
Figures



References
-
- Darwin KH, Ehrt S, Weich N, Gutierrez-Ramos J-C, Nathan CF. The proteasome of Mycobacterium tuberculosis is required for resistance to nitric oxide. Science. 2003;302:1963–1966. - PubMed
-
- Darwin KH, Lin G, Chen Z, Li H, Nathan CF. Characterization of a Mycobacterium tuberculosis proteasomal ATPase homologue. Mol Microbiol. 2005;55:561–571. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

