Role of tyrosine 33 residue for the stabilization of the tetrameric structure of human cytidine deaminase
- PMID: 20637228
- PMCID: PMC3020594
- DOI: 10.1016/j.ijbiomac.2010.07.001
Role of tyrosine 33 residue for the stabilization of the tetrameric structure of human cytidine deaminase
Erratum in
- Int J Biol Macromol. 2011 Oct 1;49(3):439
Abstract
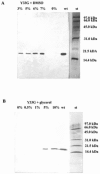
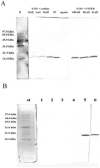
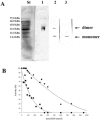
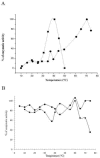
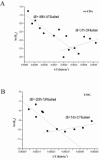
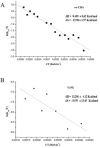
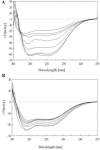
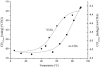
In the present work the effect of a mutation on tyrosine 33 residue (Y33G) of human cytidine deaminase (CDA) was investigated with regard to protein solubility and specific activity. Osmolytes and CDA ligands were used to increase the yield and the specific activity of the protein. The mutant enzyme was purified and subjected to a kinetic characterization and to stability studies. These investigations reinforced the hypothesis that in human CDA the side chain of Y33 is involved in intersubunit interactions with four glutamate residues (E108) forming a double latch that connects each of the two pairs of monomers of the tetrameric CDA.
Copyright © 2010 Elsevier B.V. All rights reserved.
Figures
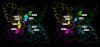
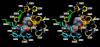
References
-
- Vincenzetti S, Quadrini B, Mariani P, De Sanctis G, Cammertoni N, Polzonetti V, Pucciarelli S, Natalini P, Vita A. Modulation of human cytidine deaminase by specific aminoacids involved in the intersubunit interactions. Proteins. 2008;70:144–156. - PubMed
-
- Costanzi S, Vincenzetti S, Cristalli G, Vita A. Human cytidine deaminase: a three-dimensional model of a tetrameric metallo-enzyme inferred from a crystal structure of a distantly related dimeric homologue. J. Mol. Graph. Model. 2006;25:10–16. - PubMed
-
- Betts L, Xiang S, Short SA, Wolfenden R, Carter CW. Cytidine deaminase. The 2.3 Ǻ crystal structure of an enzyme: transition-state analog complex. J. Mol. Biol. 1994;235:635–656. - PubMed
-
- Johansson E, Mejlhede N, Neuhard J, Larsen S. Crystal structure of the tetrameric cytidine deaminase from Bacillus subtilis at 2.0 Å resolution. Biochemistry. 2002;41:2563–2570. - PubMed
-
- Vincenzetti S, Cambi A, Maury G, Bertorelle F, Gaubert G, Neuhard J, Natalini P, Salvatori D, De Sanctis G, Vita A. Possible role of two phenylalanine residues in the active site of human cytidine deaminase. Protein Eng. 2000;13:791–799. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

