Determinants of divergent adaptation and Dobzhansky-Muller interaction in experimental yeast populations
- PMID: 20637622
- PMCID: PMC2938792
- DOI: 10.1016/j.cub.2010.06.022
Determinants of divergent adaptation and Dobzhansky-Muller interaction in experimental yeast populations
Abstract
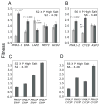
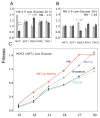
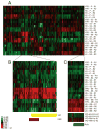
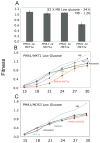
Divergent adaptation can be associated with reproductive isolation in speciation [1]. We recently demonstrated the link between divergent adaptation and the onset of reproductive isolation in experimental populations of the yeast Saccharomyces cerevisiae evolved from a single progenitor in either a high-salt or a low-glucose environment [2]. Here, whole-genome resequencing and comparative genome hybridization of representatives of three populations revealed 17 mutations, six of which explained the adaptive increases in mitotic fitness. In two populations evolved in high salt, two different mutations occurred in the proton efflux pump gene PMA1 and the global transcriptional repressor gene CYC8; the ENA genes encoding sodium efflux pumps were overexpressed once through expansion of this gene cluster and once because of mutation in the regulator CYC8. In the population from low glucose, one mutation occurred in MDS3, which modulates growth at high pH, and one in MKT1, a global regulator of mRNAs encoding mitochondrial proteins, the latter recapitulating a naturally occurring variant. A Dobzhansky-Muller (DM) incompatibility between the evolved alleles of PMA1 and MKT1 strongly depressed fitness in the low-glucose environment. This DM interaction is the first reported between experimentally evolved alleles of known genes and shows how reproductive isolation can arise rapidly when divergent selection is strong.
Copyright (c) 2010 Elsevier Ltd. All rights reserved.
Figures
References
-
- Coyne JA, Orr HA. Speciation. Sunderland: Sinauer; 2004.
-
- Dettman JR, Sirjusingh C, Kohn LM, Anderson JB. Incipient speciation by divergent adaptation and antagonistic epistasis in yeast. Nature. 2007;447:585–588. - PubMed
-
- Rundle HD, Nosil P. Ecological speciation. Ecology Letters. 2005;8:336–352.
-
- Schluter D. Evidence for ecological speciation and its alternative. Science. 2009;323:737–741. - PubMed
-
- Wu CI, Ting CT. Genes and speciation. Nat Rev Genet. 2004;5:114–122. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

