Structure based prediction of subtype-selectivity for adenosine receptor antagonists
- PMID: 20637786
- PMCID: PMC2980563
- DOI: 10.1016/j.neuropharm.2010.07.009
Structure based prediction of subtype-selectivity for adenosine receptor antagonists
Abstract
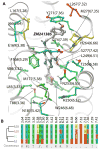
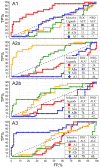
One of the major hurdles in the development of safe and effective drugs targeting G-protein coupled receptors (GPCRs) is finding ligands that are highly selective for a specific receptor subtype. Structural understanding of subtype-specific binding pocket variations and ligand-receptor interactions may greatly facilitate design of selective ligands. To gain insights into the structural basis of ligand subtype selectivity within the family of adenosine receptors (AR: A(1), A(2A), A(2B), and A(3)) we generated 3D models of all four subtypes using the recently determined crystal structure of the A(A2)AR as a template, and employing the methodology of ligand-guided receptor optimization for refinement. This approach produced 3D conformational models of AR subtypes that effectively explain binding modes and subtype selectivity for a diverse set of known AR antagonists. Analysis of the subtype-specific ligand-receptor interactions allowed identification of the major determinants of ligand selectivity, which may facilitate discovery of more efficient drug candidates.
Copyright © 2010 Elsevier Ltd. All rights reserved.
Figures



References
-
- Tyndall JD, Sandilya R. GPCR agonists and antagonists in the clinic. Med Chem. 2005;1:405–421. - PubMed
-
- Lagerstrom MC, Schioth HB. Structural diversity of G protein-coupled receptors and significance for drug discovery. Nat Rev Drug Discov. 2008;7:339–357. - PubMed
-
- Morelli M, Carta AR, Jenner P. Adenosine A(2A) Receptors and Parkinson’s Disease. Handb Exp Pharmacol. 2009:589–615. - PubMed
-
- Sebastiao AM, Ribeiro JA. Adenosine receptors and the central nervous system. Handb Exp Pharmacol. 2009:471–534. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

