Massive gene duplication event among clinical isolates of the Mycobacterium tuberculosis W/Beijing family
- PMID: 20639330
- PMCID: PMC2937425
- DOI: 10.1128/JB.00536-10
Massive gene duplication event among clinical isolates of the Mycobacterium tuberculosis W/Beijing family
Abstract
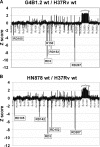
As part of our effort to uncover the molecular basis for the phenotypic variation among clinical Mycobacterium tuberculosis isolates, we have previously reported that isolates belonging to the W/Beijing lineage constitutively overexpress the DosR-regulated transcriptional program. While generating dosR knockouts in two independent W/Beijing sublineages, we were surprised to discover that they possess two copies of dosR. This dosR amplification is part of a massive genomic duplication spanning 350 kb and encompassing >300 genes. In total, this equates to 8% of the genome being present as two copies. The presence of IS6110 elements at both ends of the region of duplication, and in the novel junction region, suggests that it arose through unequal homologous recombination of sister chromatids at the IS6110 sequences. Analysis of isolates representing the major M. tuberculosis lineages has revealed that the 350-kb duplication is restricted to the most recently evolved sublineages of the W/Beijing family. Within these isolates, the duplication is partly responsible for the constitutive dosR overexpression phenotype. Although the nature of the selection event giving rise to the duplication remains unresolved, its evolution is almost certainly the result of specific selective pressure(s) encountered inside the host. A preliminary in vitro screen has failed to reveal a role of the duplication in conferring resistance to common antitubercular drugs, a trait frequently associated with W/Beijing isolates. Nevertheless, this first description of a genetic remodeling event of this nature for M. tuberculosis further highlights the potential for the evolution of diversity in this important global pathogen.
Figures

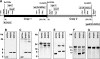


References
-
- Alland, D., D. W. Lacher, M. H. Hazbon, A. S. Motiwala, W. Qi, R. D. Fleischmann, and T. S. Whittam. 2007. Role of large sequence polymorphisms (LSPs) in generating genomic diversity among clinical isolates of Mycobacterium tuberculosis and the utility of LSPs in phylogenetic analysis. J. Clin. Microbiol. 45:39-46. - PMC - PubMed
-
- Andersson, D. I., and D. Hughes. 2009. Gene amplification and adaptive evolution in bacteria. Annu. Rev. Genet. 43:167-195. - PubMed
-
- Bifani, P. J., B. Mathema, N. E. Kurepina, and B. N. Kreiswirth. 2002. Global dissemination of the Mycobacterium tuberculosis W-Beijing family strains. Trends Microbiol. 10:45-52. - PubMed
-
- Brosch, R., S. V. Gordon, T. Garnier, K. Eiglmeier, W. Frigui, P. Valenti, S. Dos Santos, S. Duthoy, C. Lacroix, C. Garcia-Pelayo, J. K. Inwald, P. Golby, J. N. Garcia, R. G. Hewinson, M. A. Behr, M. A. Quail, C. Churcher, B. G. Barrell, J. Parkhill, and S. T. Cole. 2007. Genome plasticity of BCG and impact on vaccine efficacy. Proc. Natl. Acad. Sci. U. S. A. 104:5596-5601. - PMC - PubMed
-
- Brosch, R., S. V. Gordon, M. Marmiesse, P. Brodin, C. Buchrieser, K. Eiglmeier, T. Garnier, C. Gutierrez, G. Hewinson, K. Kremer, L. M. Parsons, A. S. Pym, S. Samper, D. van Soolingen, and S. T. Cole. 2002. A new evolutionary scenario for the Mycobacterium tuberculosis complex. Proc. Natl. Acad. Sci. U. S. A. 99:3684-3689. - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

