The prevalence of multidrug resistance is higher among bovine than human Salmonella enterica serotype Newport, Typhimurium, and 4,5,12:i:- isolates in the United States but differs by serotype and geographic region
- PMID: 20639364
- PMCID: PMC2935064
- DOI: 10.1128/AEM.00377-10
The prevalence of multidrug resistance is higher among bovine than human Salmonella enterica serotype Newport, Typhimurium, and 4,5,12:i:- isolates in the United States but differs by serotype and geographic region
Abstract
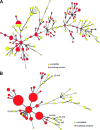
Salmonella represents an important zoonotic pathogen worldwide, but the transmission dynamics between humans and animals as well as within animal populations are incompletely understood. We characterized Salmonella isolates from cattle and humans in two geographic regions of the United States, the Pacific Northwest and the Northeast, using three common subtyping methods (pulsed-field gel electrophoresis [PFGE], multilocus variable number of tandem repeat analysis [MLVA], and multilocus sequence typing [MLST]). In addition, we analyzed the distribution of antimicrobial resistance among human and cattle Salmonella isolates from the two study areas and characterized Salmonella persistence on individual dairy farms. For both Salmonella enterica subsp. enterica serotypes Newport and Typhimurium, we found multidrug resistance to be significantly associated with bovine origin of isolates, with the odds of multidrug resistance for Newport isolates from cattle approximately 18 times higher than for Newport isolates from humans. Isolates from the Northwest were significantly more likely to be multidrug resistant than those from the Northeast, and susceptible and resistant isolates appeared to represent distinct Salmonella subtypes. We detected evidence for strain diversification during Salmonella persistence on farms, which included changes in antimicrobial resistance as well as genetic changes manifested in PFGE and MLVA pattern shifts. While discriminatory power was serotype dependent, the combination of PFGE data with either MLVA or resistance typing data consistently allowed for improved subtype discrimination. Our results are consistent with the idea that cattle are an important reservoir of multidrug-resistant Salmonella infections in humans. In addition, the study provides evidence for the value of including antimicrobial resistance data in epidemiological investigations and highlights the benefits and potential problems of combining subtyping methods.
Figures
References
-
- Alcaine, S. D., Y. Soyer, L. D. Warnick, W. L. Su, S. Sukhnanand, J. Richards, E. D. Fortes, P. McDonough, T. P. Root, N. B. Dumas, Y. Grohn, and M. Wiedmann. 2006. Multilocus sequence typing supports the hypothesis that cow- and human-associated Salmonella isolates represent distinct and overlapping populations. Appl. Environ. Microbiol. 72:7575-7585. - PMC - PubMed
-
- Alcaine, S. D., L. D. Warnick, and M. Wiedmann. 2007. Antimicrobial resistance in nontyphoidal Salmonella. J. Food Prot. 70:780-790. - PubMed
-
- Animal and Plant Inspection Service. 2005. Salmonella on U.S. dairy operations: prevalence and antimicrobial drug susceptibility. APHIS info sheet. Document N435.1005 Animal and Plant Inspection Service, Veterinary Services, Centers for Epidemiology and Animal Health, U.S. Department of Agriculture, Fort Collins, CO.
-
- Bauer, A. W., W. M. Kirby, J. C. Sherris, and M. Turck. 1966. Antibiotic susceptibility testing by a standardized single disk method. Am. J. Clin. Pathol. 45:493-496. - PubMed
-
- Belkum, A. V., P. T. Tassios, L. Dijkshoorn, S. Haeggman, B. Cookson, N. K. Fry, V. Fussing, J. Green, E. Feil, P. Gerner-Smidt, S. Brisse, and M. Struelens, and the European Society of Clinical Microbiology and Infectious Diseases (ESCMID) Study Group on Epidemiological Markers (ESGEM). 2007. Guidelines for the validation and application of typing methods for use in bacterial epidemiology. Clin. Microbiol. Infect. 13(Suppl. 3):1-46. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

