Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification
- PMID: 20639862
- PMCID: PMC3491567
- DOI: 10.1038/nature09303
Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification
Abstract
DNA methylation is one of the best-characterized epigenetic modifications. Although the enzymes that catalyse DNA methylation have been characterized, enzymes responsible for demethylation have been elusive. A recent study indicates that the human TET1 protein could catalyse the conversion of 5-methylcytosine (5mC) of DNA to 5-hydroxymethylcytosine (5hmC), raising the possibility that DNA demethylation may be a Tet1-mediated process. Here we extend this study by demonstrating that all three mouse Tet proteins (Tet1, Tet2 and Tet3) can also catalyse a similar reaction. Tet1 has an important role in mouse embryonic stem (ES) cell maintenance through maintaining the expression of Nanog in ES cells. Downregulation of Nanog via Tet1 knockdown correlates with methylation of the Nanog promoter, supporting a role for Tet1 in regulating DNA methylation status. Furthermore, knockdown of Tet1 in pre-implantation embryos results in a bias towards trophectoderm differentiation. Thus, our studies not only uncover the enzymatic activity of the Tet proteins, but also demonstrate a role for Tet1 in ES cell maintenance and inner cell mass cell specification.
Figures
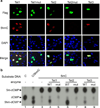
Expression of wild-type Tet proteins, but not Fe(II)-binding mutants of Tet1 or Tet2, in U2OS cells results in the generation of 5hmC. Forty-eight hours post-transfection, the cells were co-stained with Flag and 5hmC antibodies. Nuclei are counterstained by DAPI.
Recombinant catalytic domains of Tet proteins, but not their catalytic mutants, convert 5mC in DNA oligonucleotides to hmC in vitro. Double stranded DNA oligonucleotides containing a fully methylated MspI site were incubated with wild-type and catalytic mutant forms of Flag-Tet1(aa1367-2039), Flag-Tet2(916-1921), or Flag-Tet3(aa697-1668) proteins (1:10 enzyme to substrate ratio) in the presence of Fe(II) and α-KG. Recovered oligonucleotides were digested with MspI, end labeled with T4 DNA kinase, digested with DNaseI and phosphodiesterase, and analyzed by TLC. Unmethylated or 5hmC oligonucleotides containing the same sequences were used as a control for marking the migration of dCMP and hmC on TLC plates.

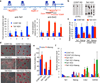
Tet1 knockdown impairs ES cell proliferation. Growth curves were determined for control and Tet1 knockdown (KD1 and KD2) cells by counting the cell numbers everyday. The average cell numbers with s.d. from three independent experiments are shown.
Tet1 knockdown impairs ES cell self-renewal. A single control or knockdown cell was plated in each well and its ability to form colonies was evaluated at day 6 after plating. There is no obvious difference between the colony size, but the colony number is greatly reduced in Tet1 knockdown cells when compared to that from control cells. Error bars represent s.d. of three independent experiments.
RT-qPCR analysis of expression levels of Tet1 and selected stem cell factors in control and knockdown cells. The expression levels in control cells are set as 1. Error bars represent s.d. of three independent experiments.
Western blot analysis of Tet1 and selected stem cell factors in control and knockdown cells. Actin is used as a loading control.
RT-qPCR analysis of the expression of various cell lineage marker genes in control and Tet1 knockdown ES cells. The expression level in control knockdown cells is set as 1. Error bars represent s.d. of three independent experiments.
ChIP analysis demonstrates that Nanog is a direct target of Tet1. Top panel is a diagram of the Nanog gene with the four amplicons indicated. The proximal and distal T-DMR as well as regions I and II are defined as that in a previous publication . The numbers in the diagram refer to the gene coordinates. Bottom left panel is Tet1 enrichment in control and Tet1 knockdown cells relative to IgG controls. Bottom right panel is 5mC enrichment in Tet1 knockdown relative to control knockdown. Results presented are the average of three independent experiments with s.d.
Bisulfite sequencing results indicate that Tet1 knockdown in ES cells results in an increase in DNA methylation at the proximal T-DMR (Fig. 3a) of Nanog promoter. Open circles indicate unmethylated CpG dinucleotides, closed circles indicate methylated CpGs.
RT-qPCR analysis demonstrates that knockdown of Tet1 in wild-type J1 ES cells, but not in the DNMT TKO J1 ES cells, results in Nanog down-regulation. Error bars represent s.d. of two independent experiments.
AP staining of mouse ES cells after puromycin selection of cells infected with lentiviruses expressing control and Tet1 knockdown shRNAs with or without co-expression of Nanog.
The self-renewal defects associated with Tet1 knockdown can be partially rescued by expression of exogenous Nanog. Self-renewal assay was performed as that described in Fig. 2b.
RT-qPCR analysis demonstrates that expression of exogenous Nanog suppressed up-regulation of differentiation genes (Cdx2, GATA6) caused by Tet1 knockdown. Data shown is the average of three independent experiments with error bars (s.e.m).
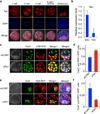
Tet1 protein is present in the nuclei of preimplantation mouse embryos. Mouse embryos at different developmental stages (one-cell to blastocyst) were stained with Tet1 antibody. Note that Tet1 is relatively enriched in the ICM when compared to that in the trophectodermal cells at blastocyst stage.
RT-qPCR demonstrates that ICM derived ES cells expressed higher levels of Tet1 when compared with TE stem cells. Error bars represent s.d. of three independent experiments.
Knockdown of Tet1 at the two-cell stage promotes trophectoderm cell specification. Representative Z-stack image of a control and a knockdown blastocyst are shown. Trophectoderm lineage cells are Cdx2 positive (green). Control knockdown and Tet1 knockdown cells are marked in red. Cells with yellow color in merge are the cells of TE lineage that were derived from the injected blastomere.
Ratio of Cdx2+RFP double positive cells over RFP positive cells from control knockdown and Tet1 knockdown. Eight control-injected embryos and 9 Tet1 siRNA injected embryos were used for the quantification analysis (p<0.001). Error bars represent s.e.m.
Knockdown of Tet1 prevents cells from contributing to ICM. Representative Z-stack images with Oct4 staining (green) in control and Tet1 knockdown cells (red). Cells with yellow color in merge are the cells derived from the injected blastomere that contributed to the ICM.
Ratio of Oct4+RFP double positive cells over RFP positive cells from control knockdown and Tet1 knockdown. Seven control-injected embryos and 7 Tet1 siRNA injected embryos were used for the quantification analysis (p=0.004). Error bars represent s.e.m.
References
-
- Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. - PubMed
-
- Cedar H, Bergman Y. Linking DNA methylation and histone modification: patterns and paradigms. Nat Rev Genet. 2009;10:295–304. - PubMed
-
- Sasaki H, Matsui Y. Epigenetic events in mammalian germ-cell development: reprogramming and beyond. Nat Rev Genet. 2008;2008:129–140. - PubMed
-
- Surani MA, Hayashi K, Hajkova P. Genetic and epigenetic regulators of pluripotency. Cell. 2007;128:747–762. - PubMed
-
- Ooi SK, Bestor TH. The colorful history of active DNA demethylation. Cell. 2008;133:1145–1148. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

