Mathematical modeling of K-Ras nanocluster formation on the plasma membrane
- PMID: 20643072
- PMCID: PMC2905126
- DOI: 10.1016/j.bpj.2010.04.055
Mathematical modeling of K-Ras nanocluster formation on the plasma membrane
Abstract
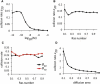
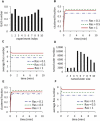
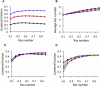
K-Ras functions as a critical node in the mitogen-activated protein kinase (MAPK) pathway that regulates key cellular functions including proliferation, differentiation, and apoptosis. Following growth factor receptor activation K-Ras.GTP forms nanoclusters on the plasma membrane through interaction with the scaffold protein galectin-3. The generation of nanoclusters is essential for high fidelity signal transduction via the MAPK pathway. To explore the mechanisms underlying K-Ras.GTP nanocluster formation, we developed a mathematical model of K-Ras-galectin-3 interactions. We designed a computational method to calculate protein collision rates based on experimentally determined protein diffusion rates and diffusion mechanisms and used a genetic algorithm to search the values of key model parameters. The optimal estimated model parameters were validated using experimental data. The resulting model accurately replicates critical features of K-Ras nanoclustering, including a fixed ratio of clustered K-Ras.GTP to monomeric K-Ras.GTP that is independent of the concentration of K-Ras.GTP. The model reproduces experimental results showing that the cytosolic level of galectin-3 determines the magnitude of the K-Ras.GTP clustered fraction and illustrates that nanoclustering is regulated by key nonequilibrium processes. Our kinetic model identifies a potential biophysical mechanism for K-Ras nanoclustering and suggests general principles that may be relevant for other plasma-membrane-localized proteins.
Copyright (c) 2010 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures


References
-
- Cox A.D., Der C.J. The dark side of Ras: regulation of apoptosis. Oncogene. 2003;22:8999–9006. - PubMed
-
- Downward J. Targeting RAS signalling pathways in cancer therapy. Nat. Rev. Cancer. 2003;3:11–22. - PubMed
-
- Bhalla U.S., Ram P.T., Iyengar R. MAP kinase phosphatase as a locus of flexibility in a mitogen-activated protein kinase signaling network. Science. 2002;297:1018–1023. - PubMed
-
- Tian T., Harding A., Hancock J.F. Plasma membrane nanoswitches generate high-fidelity Ras signal transduction. Nat. Cell Biol. 2007;9:905–914. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

