Structural evidence that peroxiredoxin catalytic power is based on transition-state stabilization
- PMID: 20643143
- PMCID: PMC2941395
- DOI: 10.1016/j.jmb.2010.07.022
Structural evidence that peroxiredoxin catalytic power is based on transition-state stabilization
Abstract
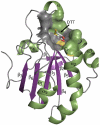
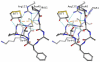
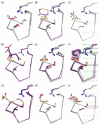
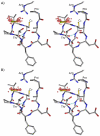
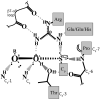
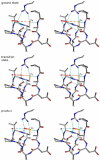
Peroxiredoxins (Prxs) are important peroxidases associated with both antioxidant protection and redox signaling. They use a conserved Cys residue to reduce peroxide substrates. The Prxs have a remarkably high catalytic efficiency that makes them a dominant player in cell-wide peroxide reduction, but the origins of their high activity have been mysterious. We present here a novel structure of human PrxV at 1.45 A resolution that has a dithiothreitol bound in the active site with its diol moiety mimicking the two oxygens of a peroxide substrate. This suggests diols and similar di-oxygen compounds as a novel class of competitive inhibitors for the Prxs. Common features of this and other structures containing peroxide, peroxide-mimicking ligands, or peroxide-mimicking water molecules reveal hydrogen bonding and steric factors that promote its high reactivity by creating an oxygen track along which the peroxide oxygens move as the reaction proceeds. Key insights include how the active-site microenvironment activates both the peroxidatic cysteine side chain and the peroxide substrate and how it is exquisitely well suited to stabilize the transition state of the in-line S(N)2 substitution reaction that is peroxidation.
Copyright 2010 Elsevier Ltd. All rights reserved.
Figures
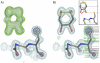
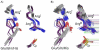
References
-
- Karplus PA, Hall A. Structural survey of the peroxiredoxins. In: Flohé L, Harris JR, editors. Peroxiredoxin Systems. Springer; New York: 2007. pp. 41–60. - PubMed
-
- Flohe L, Budde H, Hofmann B. Peroxiredoxins in antioxidant defense and redox regulation. Biofactors. 2003;19:3–10. - PubMed
-
- Akerman SE, Muller S. Peroxiredoxin-linked detoxification of hydroperoxides in Toxoplasma gondii. J Biol Chem. 2005;280:564–70. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

