Envelope glycans of immunodeficiency virions are almost entirely oligomannose antigens
- PMID: 20643940
- PMCID: PMC2922250
- DOI: 10.1073/pnas.1006498107
Envelope glycans of immunodeficiency virions are almost entirely oligomannose antigens
Abstract
The envelope spike of HIV is one of the most highly N-glycosylated structures found in nature. However, despite extensive research revealing essential functional roles in infection and immune evasion, the chemical structures of the glycans on the native viral envelope glycoprotein gp120--as opposed to recombinantly generated gp120--have not been described. Here, we report on the identity of the N-linked glycans from primary isolates of HIV-1 (clades A, B, and C) and from the simian immunodeficiency virus. MS analysis reveals a remarkably simple and highly conserved virus-specific glycan profile almost entirely devoid of medial Golgi-mediated processing. In stark contrast to recombinant gp120, which shows extensive exposure to cellular glycosylation enzymes (>70% complex type glycans), the native envelope shows barely detectable processing beyond the biosynthetic intermediate Man5GlcNAc2 (<2% complex type glycans). This oligomannose (Man5-9GlcNAc2) profile is conserved across primary isolates and geographically divergent clades but is not reflected in the current generation of gp120 antigens used for vaccine trials. In the context of vaccine design, we also note that Manalpha1-->2Man-terminating glycans (Man6-9GlcNAc2) of the type recognized by the broadly neutralizing anti-HIV antibody 2G12 are 3-fold more abundant on the native envelope than on the recombinant monomer and are also found on isolates not neutralized by 2G12. The Manalpha1-->2Man residues of gp120 therefore provide a vaccine target that is physically larger and antigenically more conserved than the 2G12 epitope itself. This study revises and extends our understanding of the glycan shield of HIV with implications for AIDS vaccine design.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

 , Fuc; ⋆, NeuNAc (50). The linkage position is shown by the angle of the lines linking the sugar residues (vertical line, two-link; forward slash, three-link; horizontal line, four-link; back slash, six-link). Anomericity is indicated by full lines for β-bonds and by broken lines for α-bonds (50).
, Fuc; ⋆, NeuNAc (50). The linkage position is shown by the angle of the lines linking the sugar residues (vertical line, two-link; forward slash, three-link; horizontal line, four-link; back slash, six-link). Anomericity is indicated by full lines for β-bonds and by broken lines for α-bonds (50).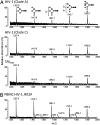


References
-
- Zhu X, Borchers C, Bienstock RJ, Tomer KB. Mass spectrometric characterization of the glycosylation pattern of HIV-gp120 expressed in CHO cells. Biochemistry. 2000;39:11194–11204. - PubMed
-
- Pantophlet R, Burton DR. GP120: Target for neutralizing HIV-1 antibodies. Annu Rev Immunol. 2006;24:739–769. - PubMed
-
- Wei X, et al. Antibody neutralization and escape by HIV-1. Nature. 2003;422:307–312. - PubMed
-
- Scanlan CN, Offer J, Zitzmann N, Dwek RA. Exploiting the defensive sugars of HIV-1 for drug and vaccine design. Nature. 2007;446:1038–1045. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

