Active RNA polymerases: mobile or immobile molecular machines?
- PMID: 20644712
- PMCID: PMC2903595
- DOI: 10.1371/journal.pbio.1000419
Active RNA polymerases: mobile or immobile molecular machines?
Abstract
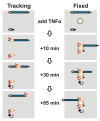
It is widely assumed that active RNA polymerases track along their templates to produce a transcript. We test this using chromosome conformation capture and human genes switched on rapidly and synchronously by tumour necrosis factor alpha (TNFalpha); one is 221 kbp SAMD4A, which a polymerase takes more than 1 h to transcribe. Ten minutes after stimulation, the SAMD4A promoter comes together with other TNFalpha-responsive promoters. Subsequently, these contacts are lost as new downstream ones appear; contacts are invariably between sequences being transcribed. Super-resolution microscopy confirms that nascent transcripts (detected by RNA fluorescence in situ hybridization) co-localize at relevant times. Results are consistent with an alternative view of transcription: polymerases fixed in factories reel in their respective templates, so different parts of the templates transiently lie together.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




Similar articles
-
TNFα signals through specialized factories where responsive coding and miRNA genes are transcribed.EMBO J. 2012 Nov 28;31(23):4404-14. doi: 10.1038/emboj.2012.288. Epub 2012 Oct 26. EMBO J. 2012. PMID: 23103767 Free PMC article.
-
Dynamic reconfiguration of long human genes during one transcription cycle.Mol Cell Biol. 2012 Jul;32(14):2738-47. doi: 10.1128/MCB.00179-12. Epub 2012 May 14. Mol Cell Biol. 2012. PMID: 22586273 Free PMC article.
-
Space exploration by the promoter of a long human gene during one transcription cycle.Nucleic Acids Res. 2013 Feb 1;41(4):2216-27. doi: 10.1093/nar/gks1441. Epub 2013 Jan 8. Nucleic Acids Res. 2013. PMID: 23303786 Free PMC article.
-
The organization of replication and transcription.Science. 1999 Jun 11;284(5421):1790-5. doi: 10.1126/science.284.5421.1790. Science. 1999. PMID: 10364545 Review.
-
Transcription activation and repression by interaction of a regulator with the alpha subunit of RNA polymerase: the model of phage phi 29 protein p4.Prog Nucleic Acid Res Mol Biol. 1998;60:29-46. doi: 10.1016/s0079-6603(08)60888-0. Prog Nucleic Acid Res Mol Biol. 1998. PMID: 9594570 Review.
Cited by
-
Super-Resolution Microscopy in Studying the Structure and Function of the Cell Nucleus.Acta Naturae. 2017 Oct-Dec;9(4):42-51. Acta Naturae. 2017. PMID: 29340216 Free PMC article.
-
TNFα signals through specialized factories where responsive coding and miRNA genes are transcribed.EMBO J. 2012 Nov 28;31(23):4404-14. doi: 10.1038/emboj.2012.288. Epub 2012 Oct 26. EMBO J. 2012. PMID: 23103767 Free PMC article.
-
Heat shock reduces stalled RNA polymerase II and nucleosome turnover genome-wide.Genes Dev. 2011 Nov 15;25(22):2387-97. doi: 10.1101/gad.177675.111. Genes Dev. 2011. PMID: 22085965 Free PMC article.
-
An emerging paradigm in epigenetic marking: coordination of transcription and replication.Transcription. 2024 Feb-Apr;15(1-2):22-37. doi: 10.1080/21541264.2024.2316965. Epub 2024 Feb 20. Transcription. 2024. PMID: 38378467 Free PMC article. Review.
-
Transcription and recombination factories: common features?Curr Opin Cell Biol. 2011 Jun;23(3):318-24. doi: 10.1016/j.ceb.2010.11.007. Epub 2010 Dec 17. Curr Opin Cell Biol. 2011. PMID: 21169003 Free PMC article. Review.
References
-
- Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P. Molecular biology of the cell 4th edition. New York: Garland Science; 2002. 1463
-
- Jackson D. A, McCready S. J, Cook P. R. RNA is synthesised at the nuclear cage. Nature. 1981;292:552–555. - PubMed
-
- Cook P. R. The organization of replication and transcription. Science. 1999;284:1790–1795. - PubMed
-
- Sexton T, Umlauf D, Kurukuti S, Fraser P. The role of transcription factories in large-scale structure and dynamics of interphase chromatin. Semin Cell Dev Biol. 2007;18:691–697. - PubMed
-
- Cook P. R. A model for all genomes; the role of transcription factories. J Mol Biol. 2009;395:1–10. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

