Deciphering ionic current signatures of DNA transport through a nanopore
- PMID: 20644747
- PMCID: PMC2909628
- DOI: 10.1039/b9nr00275h
Deciphering ionic current signatures of DNA transport through a nanopore
Abstract
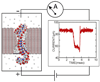
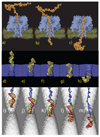
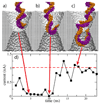
Within just a decade from the pioneering work demonstrating the utility of nanopores for molecular sensing, nanopores have emerged as versatile systems for single-molecule manipulation and analysis. In a typical setup, a gradient of the electrostatic potential captures charged solutes from the solution and forces them to move through a single nanopore, across an otherwise impermeable membrane. The ionic current blockades resulting from the presence of a solute in a nanopore can reveal the type of the solute, for example, the nucleotide makeup of a DNA strand. Despite great success, the microscopic mechanisms underlying the functionality of such stochastic sensors remain largely unknown, as it is not currently possible to characterize the microscopic conformations of single biomolecules directly in a nanopore and thereby unequivocally establish the causal relationship between the observables and the microscopic events. Such a relationship can be determined using molecular dynamics-a computational method that can accurately predict the time evolution of a molecular system starting from a given microscopic state. This article describes recent applications of this method to the process of DNA transport through biological and synthetic nanopores.
Figures





References
-
- Dekker C. Solid-state nanopores. Nature Nanotech. 2007;2:209–215. - PubMed
-
- Storm AJ, Chen JH, Zandbergen HW, Dekker C. Translocation of double-strand DNA through a silicon oxide nanopore. Phys. Rev. E. 2005;71:051903–051913. - PubMed
-
- Hornblower B, Coombs A, Whitaker RD, Kolomeisky A, Picone SJ, Meller A, Akeson M. Single-molecule analysis of DNA-protein complexes using nanopores. Nature Mater. 2007;4:315–317. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

