The ANISEED database: digital representation, formalization, and elucidation of a chordate developmental program
- PMID: 20647237
- PMCID: PMC2945195
- DOI: 10.1101/gr.108175.110
The ANISEED database: digital representation, formalization, and elucidation of a chordate developmental program
Abstract
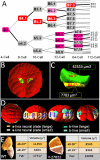
Developmental biology aims to understand how the dynamics of embryonic shapes and organ functions are encoded in linear DNA molecules. Thanks to recent progress in genomics and imaging technologies, systemic approaches are now used in parallel with small-scale studies to establish links between genomic information and phenotypes, often described at the subcellular level. Current model organism databases, however, do not integrate heterogeneous data sets at different scales into a global view of the developmental program. Here, we present a novel, generic digital system, NISEED, and its implementation, ANISEED, to ascidians, which are invertebrate chordates suitable for developmental systems biology approaches. ANISEED hosts an unprecedented combination of anatomical and molecular data on ascidian development. This includes the first detailed anatomical ontologies for these embryos, and quantitative geometrical descriptions of developing cells obtained from reconstructed three-dimensional (3D) embryos up to the gastrula stages. Fully annotated gene model sets are linked to 30,000 high-resolution spatial gene expression patterns in wild-type and experimentally manipulated conditions and to 528 experimentally validated cis-regulatory regions imported from specialized databases or extracted from 160 literature articles. This highly structured data set can be explored via a Developmental Browser, a Genome Browser, and a 3D Virtual Embryo module. We show how integration of heterogeneous data in ANISEED can provide a system-level understanding of the developmental program through the automatic inference of gene regulatory interactions, the identification of inducing signals, and the discovery and explanation of novel asymmetric divisions.
Figures




References
-
- Audic S, Claverie JM 1997. The significance of digital gene expression profiles. Genome Res 7: 986–995 - PubMed
-
- Bertrand V, Hudson C, Caillol D, Popovici C, Lemaire P 2003. Neural tissue in ascidian embryos is induced by FGF9/16/20, acting via a combination of maternal GATA and Ets transcription factors. Cell 115: 615–627 - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
