The Drosophila SUN protein Spag4 cooperates with the coiled-coil protein Yuri Gagarin to maintain association of the basal body and spermatid nucleus
- PMID: 20647369
- PMCID: PMC2915878
- DOI: 10.1242/jcs.066589
The Drosophila SUN protein Spag4 cooperates with the coiled-coil protein Yuri Gagarin to maintain association of the basal body and spermatid nucleus
Abstract
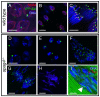
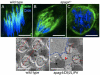
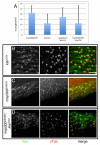
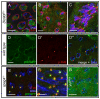
Maintaining the proximity of centrosomes to nuclei is important in several cellular contexts, and LINC complexes formed by SUN and KASH proteins are crucial in this process. Here, we characterize the presumed Drosophila ortholog of the mammalian SUN protein, sperm-associated antigen 4 (Spag4, previously named Giacomo), and demonstrate that Spag4 is required for centriole and nuclear attachment during spermatogenesis. Production of spag4 mRNA is limited to the testis, and Spag4 protein shows a dynamic pattern of association with the germline nuclei, including a concentration of protein at the site of attachment of the single spermatid centriole. In the absence of Spag4, nuclei and centrioles or basal bodies (BBs) dissociate from each other after meiosis. This role of Spag4 in centriolar attachment does not involve either of the two KASH proteins of the Drosophila genome (Klarsicht and MSP-300), but does require the coiled-coil protein Yuri Gagarin. Yuri shows an identical pattern of localization at the nuclear surface to Spag4 during spermatogenesis, and epistasis studies show that the activities of Yuri and dynein-dynactin are downstream of spag4 in this centriole attachment pathway. The later defects in spermatogenesis seen for yuri and spag4 mutants are similar, suggesting they could be secondary to initial disruption of events at the nuclear surface.
Figures



Similar articles
-
Yuri gagarin is required for actin, tubulin and basal body functions in Drosophila spermatogenesis.J Cell Sci. 2008 Jun 1;121(11):1926-36. doi: 10.1242/jcs.026559. Epub 2008 May 13. J Cell Sci. 2008. PMID: 18477609
-
Asunder is a critical regulator of dynein-dynactin localization during Drosophila spermatogenesis.Mol Biol Cell. 2009 Jun;20(11):2709-21. doi: 10.1091/mbc.e08-12-1165. Epub 2009 Apr 8. Mol Biol Cell. 2009. PMID: 19357193 Free PMC article.
-
Regulation of dynein localization and centrosome positioning by Lis-1 and asunder during Drosophila spermatogenesis.Development. 2012 Aug;139(16):2945-54. doi: 10.1242/dev.077511. Epub 2012 Jul 4. Development. 2012. PMID: 22764052 Free PMC article.
-
SAPs as a new model to probe the pathway of centriole and centrosome assembly.Biochem Soc Trans. 2021 Jun 30;49(3):1233-1240. doi: 10.1042/BST20200833. Biochem Soc Trans. 2021. PMID: 33960367 Free PMC article. Review.
-
Role of DZIP1-CBY-FAM92 transition zone complex in the basal body to membrane attachment and ciliary budding.Biochem Soc Trans. 2020 Jun 30;48(3):1067-1075. doi: 10.1042/BST20191007. Biochem Soc Trans. 2020. PMID: 32491167 Review.
Cited by
-
Bivalent individualization during chromosome territory formation in Drosophila spermatocytes by controlled condensin II protein activity and additional force generators.PLoS Genet. 2021 Oct 20;17(10):e1009870. doi: 10.1371/journal.pgen.1009870. eCollection 2021 Oct. PLoS Genet. 2021. PMID: 34669718 Free PMC article.
-
Drosophila Pif1A is essential for spermatogenesis and is the homolog of human CCDC157, a gene associated with idiopathic NOA.Cell Death Dis. 2019 Feb 11;10(2):125. doi: 10.1038/s41419-019-1398-3. Cell Death Dis. 2019. PMID: 30741974 Free PMC article.
-
The proximal centriole-like structure maintains nucleus-centriole architecture in sperm.J Cell Sci. 2024 Sep 1;137(17):jcs262311. doi: 10.1242/jcs.262311. Epub 2024 Sep 6. J Cell Sci. 2024. PMID: 39166297 Free PMC article.
-
An orphan gene is essential for efficient sperm entry into eggs in Drosophila melanogaster.bioRxiv [Preprint]. 2024 Dec 14:2024.08.08.607187. doi: 10.1101/2024.08.08.607187. bioRxiv. 2024. Update in: Genetics. 2025 Mar 17;229(3):iyaf008. doi: 10.1093/genetics/iyaf008. PMID: 39149251 Free PMC article. Updated. Preprint.
-
A deficiency screen identifies genomic regions critical for sperm head-tail connection.G3 (Bethesda). 2025 Feb 5;15(2):jkae275. doi: 10.1093/g3journal/jkae275. G3 (Bethesda). 2025. PMID: 39700389 Free PMC article.
References
-
- Arama E., Steller H. (2006). Detection of apoptosis by terminal deoxynucleotidyl transferase-mediated dUTP nick-end labeling and acridine orange in Drosophila embryos and adult male gonads. Nat. Protoc. 1, 1725-1731 - PubMed
-
- Arama E., Agapite J., Steller H. (2003). Caspase activity and a specific cytochrome C are required for sperm differentiation in Drosophila. Dev. Cell 4, 687-697 - PubMed
-
- Armstrong J. D., Texada M. J., Munjaal R., Baker D. A., Beckingham K. M. (2006). Gravitaxis in Drosophila melanogaster: a forward genetic screen. Genes Brain Behav. 5, 222-239 - PubMed
-
- Bray J. D., Chennathukuzhi V. M., Hecht N. B. (2002). Identification and characterization of cDNAs encoding four novel proteins that interact with translin associated factor-X. Genomics 79, 799-808 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

