Quantitative proteomic analysis of A549 cells infected with human respiratory syncytial virus
- PMID: 20647383
- PMCID: PMC2984239
- DOI: 10.1074/mcp.M110.001859
Quantitative proteomic analysis of A549 cells infected with human respiratory syncytial virus
Abstract
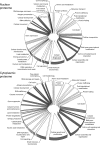
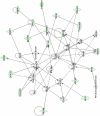
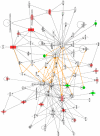
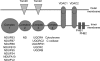
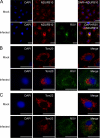
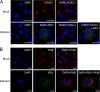
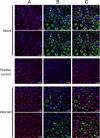
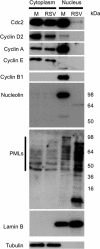
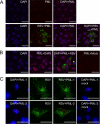
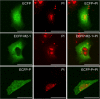
Human respiratory syncytial virus (HRSV) is a major cause of pediatric lower respiratory tract disease to which there is no vaccine or efficacious chemotherapeutic strategy. Although RNA synthesis and virus assembly occur in the cytoplasm, HRSV is known to induce nuclear responses in the host cell as replication alters global gene expression. Quantitative proteomics was used to take an unbiased overview of the protein changes in transformed human alveolar basal epithelial cells infected with HRSV. Underpinning this was the use of stable isotope labeling with amino acids in cell culture coupled to LC-MS/MS, which allowed the direct and simultaneous identification and quantification of both cellular and viral proteins. To reduce sample complexity and increase data return on potential protein localization, cells were fractionated into nuclear and cytoplasmic extracts. This resulted in the identification of 1,140 cellular proteins and six viral proteins. The proteomics data were analyzed using Ingenuity Pathways Analysis to identify defined canonical pathways and functional groupings. Selected data were validated using Western blot, direct and indirect immunofluorescence confocal microscopy, and functional assays. The study served to validate and expand upon known HRSV-host cell interactions, including those associated with the antiviral response and alterations in subnuclear structures such as the nucleolus and ND10 (promyelocytic leukemia bodies). In addition, novel changes were observed in mitochondrial proteins and functions, cell cycle regulatory molecules, nuclear pore complex proteins and nucleocytoplasmic trafficking proteins. These data shed light into how the cell is potentially altered to create conditions more favorable for infection. Additionally, the study highlights the application and advantage of stable isotope labeling with amino acids in cell culture coupled to LC-MS/MS for the analysis of virus-host interactions.
Figures



References
-
- Openshaw P. J., Dean G. S., Culley F. J. (2003) Links between respiratory syncytial virus bronchiolitis and childhood asthma: clinical and research approaches. Pediatr. Infect. Dis. J. 22, S58–S64; discussion S64–S65 - PubMed
-
- Martinez F. D. (2003) Respiratory syncytial virus bronchiolitis and the pathogenesis of childhood asthma. Pediatr. Infect. Dis. J. 22, S76–82 - PubMed
-
- Collins P. L., Murphy B. R. (2002) Respiratory syncytial virus: reverse genetics and vaccine strategies. Virology 296, 204–211 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

