Las1L is a nucleolar protein required for cell proliferation and ribosome biogenesis
- PMID: 20647540
- PMCID: PMC2937536
- DOI: 10.1128/MCB.00358-10
Las1L is a nucleolar protein required for cell proliferation and ribosome biogenesis
Abstract
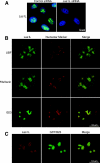
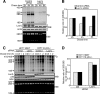
Ribosome biogenesis is a highly regulated process ensuring that cell growth (increase in biomass) is coordinated with cell proliferation. The formation of eukaryotic ribosomes is a multistep process initiated by the transcription and processing of rRNA in the nucleolus. Concomitant with this, several preribosomal particles, which transiently associate with numerous nonribosomal factors before mature 60S and 40S subunits are formed and exported in the cytoplasm, are generated. Here we identify Las1L as a previously uncharacterized nucleolar protein required for ribosome biogenesis. Depletion of Las1L causes inhibition of cell proliferation characterized by a G1 arrest dependent on the tumor suppressor p53. Moreover, we demonstrate that Las1L is crucial for ribosome biogenesis and that depletion of Las1L leads to inhibition of rRNA processing and failure to synthesize the mature 28S rRNA. Taken together, our data demonstrate that Las1L is essential for cell proliferation and biogenesis of the 60S ribosomal subunit.
Figures






References
-
- Arabi, A., S. Wu, K. Ridderstrale, H. Bierhoff, C. Shiue, K. Fatyol, S. Fahlen, P. Hydbring, O. Soderberg, I. Grummt, L. G. Larsson, and A. P. Wright. 2005. c-Myc associates with ribosomal DNA and activates RNA polymerase I transcription. Nat. Cell Biol. 7:303-310. - PubMed
-
- Belin, S., A. Beghin, E. Solano-Gonzalez, L. Bezin, S. Brunet-Manquat, J. Textoris, A. C. Prats, H. C. Mertani, C. Dumontet, and J. J. Diaz. 2009. Dysregulation of ribosome biogenesis and translational capacity is associated with tumor progression of human breast cancer cells. PLoS One 4:e7147. - PMC - PubMed
-
- Boisvert, F. M., S. van Koningsbruggen, J. Navascues, and A. I. Lamond. 2007. The multifunctional nucleolus. Nat. Rev. Mol. Cell Biol. 8:574-585. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous
