Severe Progressive Autism Associated with Two de novo Changes: A 2.6-Mb 2q31.1 Deletion and a Balanced t(14;21)(q21.1;p11.2) Translocation with Long-Range Epigenetic Silencing of LRFN5 Expression
- PMID: 20648246
- PMCID: PMC2883852
- DOI: 10.1159/000280290
Severe Progressive Autism Associated with Two de novo Changes: A 2.6-Mb 2q31.1 Deletion and a Balanced t(14;21)(q21.1;p11.2) Translocation with Long-Range Epigenetic Silencing of LRFN5 Expression
Abstract
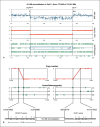
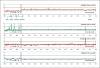
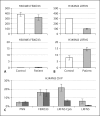
In a 19-year-old severely autistic and mentally retarded girl, a balanced de novo t(14;21)(q21.1;p11.2) translocation was found in addition to a de novo 2.6-Mb 2q31.1 deletion containing 15 protein-encoding genes. To investigate if the translocation might contribute to developmental stagnation at the age of 2 years with later regression of skills, i.e. a more severe phenotype than expected from the 2q31.1 deletion, the epigenetic status and expression of genes proximal and distal to the 14q21.1 breakpoint were investigated in Ebstein Barr Virus-transformed lymphoblast and primary skin fibroblast cells. The 14q21.1 breakpoint was found to be located between a cluster of 7 genes 0.1 Mb upstream, starting with FBXO33, and the single and isolated LRFN5 gene 2.1 Mb downstream. Only expression of LRFN5 appeared to be affected by its novel genomic context. In patient fibroblasts, LRFN5 expression was 10-fold reduced compared to LRFN5 expressed in control fibroblasts. In addition, a relative increase in trimethylated histone H3 lysine 9 (H3K9M3)-associated DNA starting exactly at the translocation breakpoint and going 2.5 Mb beyond the LRFN5 gene was found. At the LRFN5 promoter, there was a distinct peak of trimethylated histone H3 lysine 27 (H3K27M3)-associated DNA in addition to a diminished trimethylated histone H3 lysine 4 (H3K4M3) level. We speculate that dysregulation of LRFN5, a postsynaptic density-associated gene, may contribute to the patient's autism, even though 2 other patients with 14q13.2q21.3 deletions that included LRFN5 were not autistic. More significantly, we have shown that translocations may influence gene expression more than 2 Mb away from the translocation breakpoint.
Keywords: Deletion 2q31.1; Epigenetics; Gene silencing; LRFN5; Translocation.
Figures


Similar articles
-
LRFN5 locus structure is associated with autism and influenced by the sex of the individual and locus conversions.Autism Res. 2022 Mar;15(3):421-433. doi: 10.1002/aur.2677. Epub 2022 Jan 28. Autism Res. 2022. PMID: 35088940 Free PMC article.
-
Deciphering complex rearrangements at the breakpoint of an apparently balanced reciprocal translocation t(4:18)(q31;q11.2)dn and at a cryptic deletion: Further evidence of TLL1 as a causative gene for atrial septal defect.Am J Med Genet A. 2022 Aug;188(8):2472-2478. doi: 10.1002/ajmg.a.62777. Epub 2022 May 14. Am J Med Genet A. 2022. PMID: 35567499
-
Cryptic genomic imbalances in patients with de novo or familial apparently balanced translocations and abnormal phenotype.Mol Cytogenet. 2008 Jul 21;1:15. doi: 10.1186/1755-8166-1-15. Mol Cytogenet. 2008. PMID: 18644119 Free PMC article.
-
Human AZFb deletions cause distinct testicular pathologies depending on their extensions in Yq11 and the Y haplogroup: new cases and review of literature.Cell Biosci. 2021 Mar 25;11(1):60. doi: 10.1186/s13578-021-00551-2. Cell Biosci. 2021. PMID: 33766143 Free PMC article. Review.
-
Central nervous system abnormalities and psychomotor retardation in a girl with a 15.4-MB deletion of 14q12→q21.2 and a 550-KB deletion of 18p11.23: microarray delineation of an unbalanced chromosome rearrangement and a literature review.Genet Couns. 2016;27(2):165-76. Genet Couns. 2016. PMID: 29485807 Review.
Cited by
-
Proper migration and axon outgrowth of zebrafish cranial motoneuron subpopulations require the cell adhesion molecule MDGA2A.Biol Open. 2015 Jan 8;4(2):146-54. doi: 10.1242/bio.20148482. Biol Open. 2015. PMID: 25572423 Free PMC article.
-
LRFN5 locus structure is associated with autism and influenced by the sex of the individual and locus conversions.Autism Res. 2022 Mar;15(3):421-433. doi: 10.1002/aur.2677. Epub 2022 Jan 28. Autism Res. 2022. PMID: 35088940 Free PMC article.
-
Case-control genome-wide association study of persistent attention-deficit hyperactivity disorder identifies FBXO33 as a novel susceptibility gene for the disorder.Neuropsychopharmacology. 2015 Mar;40(4):915-26. doi: 10.1038/npp.2014.267. Epub 2014 Oct 6. Neuropsychopharmacology. 2015. PMID: 25284319 Free PMC article.
-
Heterozygous deletion of the LRFN2 gene is associated with working memory deficits.Eur J Hum Genet. 2016 Jun;24(6):911-8. doi: 10.1038/ejhg.2015.221. Epub 2015 Oct 21. Eur J Hum Genet. 2016. PMID: 26486473 Free PMC article.
-
LRFN5 and OLFM4 as novel potential biomarkers for major depressive disorder: a pilot study.Transl Psychiatry. 2023 Jun 6;13(1):188. doi: 10.1038/s41398-023-02490-7. Transl Psychiatry. 2023. PMID: 37280213 Free PMC article.
References
-
- Barber JC, Zhang S, Friend N, Collins AL, Maloney VK, et al. Duplications of proximal 16q flanked by heterochromatin are not euchromatic variants and show no evidence of heterochromatic position effect. Cytogenet Genome Res. 2006;114:351–358. - PubMed
-
- Clark SJ. Action at a distance: epigenetic silencing of large chromosomal regions in carcinogenesis. Hum Mol Genet. 2007;16(Spec No 1):R88–R95. - PubMed
-
- Cremer T, Cremer C. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat Rev Genet. 2001;2:292–301. - PubMed
-
- de Bruijn DR, Kater-Baats E, Eleveld M, Merkx G, Geurts Van Kessel A. Mapping and characterization of the mouse and human SS18 genes, two human SS18-like genes and a mouse Ss18 pseudogene. Cytogenet Cell Genet. 2001;92:310–319. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

