The major determinant of exendin-4/glucagon-like peptide 1 differential affinity at the rat glucagon-like peptide 1 receptor N-terminal domain is a hydrogen bond from SER-32 of exendin-4
- PMID: 20649595
- PMCID: PMC2958643
- DOI: 10.1111/j.1476-5381.2010.00834.x
The major determinant of exendin-4/glucagon-like peptide 1 differential affinity at the rat glucagon-like peptide 1 receptor N-terminal domain is a hydrogen bond from SER-32 of exendin-4
Abstract
Background and purpose: Exendin-4 (exenatide, Ex4) is a high-affinity peptide agonist at the glucagon-like peptide-1 receptor (GLP-1R), which has been approved as a treatment for type 2 diabetes. Part of the drug/hormone binding site was described in the crystal structures of both GLP-1 and Ex4 bound to the isolated N-terminal domain (NTD) of GLP-1R. However, these structures do not account for the large difference in affinity between GLP-1 and Ex4 at this isolated domain, or for the published role of the C-terminal extension of Ex4. Our aim was to clarify the pharmacology of GLP-1R in the context of these new structural data.
Experimental approach: The affinities of GLP-1, Ex4 and various analogues were measured at human and rat GLP-1R (hGLP-1R and rGLP-1R, respectively) and various receptor variants. Molecular dynamics coupled with in silico mutagenesis were used to model and interpret the data.
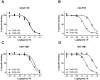
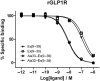
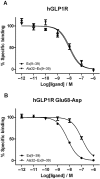
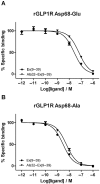
Key results: The membrane-tethered NTD of hGLP-1R displayed similar affinity for GLP-1 and Ex4 in sharp contrast to previous studies using the soluble isolated domain. The selectivity at rGLP-1R for Ex4(9-39) over Ex4(9-30) was due to Ser-32 in the ligand. While this selectivity was not observed at hGLP-1R, it was regained when Glu-68 of hGLP-1R was mutated to Asp.
Conclusions and implications: GLP-1 and Ex4 bind to the NTD of hGLP-1R with similar affinity. A hydrogen bond between Ser32 of Ex4 and Asp-68 of rGLP-1R, which is not formed with Glu-68 of hGLP-1R, is responsible for the improved affinity of Ex4 at the rat receptor.
Figures
Similar articles
-
Differential structural properties of GLP-1 and exendin-4 determine their relative affinity for the GLP-1 receptor N-terminal extracellular domain.Biochemistry. 2007 May 15;46(19):5830-40. doi: 10.1021/bi062309m. Epub 2007 Apr 20. Biochemistry. 2007. PMID: 17444618
-
Residues within the transmembrane domain of the glucagon-like peptide-1 receptor involved in ligand binding and receptor activation: modelling the ligand-bound receptor.Mol Endocrinol. 2011 Oct;25(10):1804-18. doi: 10.1210/me.2011-1160. Epub 2011 Aug 25. Mol Endocrinol. 2011. PMID: 21868452 Free PMC article.
-
Glucagon-like peptide-1 receptor ligand interactions: structural cross talk between ligands and the extracellular domain.PLoS One. 2014 Sep 2;9(9):e105683. doi: 10.1371/journal.pone.0105683. eCollection 2014. PLoS One. 2014. PMID: 25180755 Free PMC article.
-
Pharmacology of exenatide (synthetic exendin-4): a potential therapeutic for improved glycemic control of type 2 diabetes.Regul Pept. 2004 Feb 15;117(2):77-88. doi: 10.1016/j.regpep.2003.10.028. Regul Pept. 2004. PMID: 14700743 Review.
-
The structure and function of the glucagon-like peptide-1 receptor and its ligands.Br J Pharmacol. 2012 May;166(1):27-41. doi: 10.1111/j.1476-5381.2011.01687.x. Br J Pharmacol. 2012. PMID: 21950636 Free PMC article. Review.
Cited by
-
Physiology and emerging biochemistry of the glucagon-like peptide-1 receptor.Exp Diabetes Res. 2012;2012:470851. doi: 10.1155/2012/470851. Epub 2012 May 14. Exp Diabetes Res. 2012. PMID: 22666230 Free PMC article. Review.
-
Purification of family B G protein-coupled receptors using nanodiscs: Application to human glucagon-like peptide-1 receptor.PLoS One. 2017 Jun 13;12(6):e0179568. doi: 10.1371/journal.pone.0179568. eCollection 2017. PLoS One. 2017. PMID: 28609478 Free PMC article.
-
Exendin-4 as a Versatile Therapeutic Agent for the Amelioration of Diabetic Changes.Adv Pharm Bull. 2022 Mar;12(2):237-247. doi: 10.34172/apb.2022.025. Epub 2021 Jan 31. Adv Pharm Bull. 2022. PMID: 35620334 Free PMC article. Review.
-
The effect of acylation with fatty acids and other modifications on HLA class II:peptide binding and T cell stimulation for three model peptides.PLoS One. 2018 May 14;13(5):e0197407. doi: 10.1371/journal.pone.0197407. eCollection 2018. PLoS One. 2018. PMID: 29758051 Free PMC article.
-
Second extracellular loop of human glucagon-like peptide-1 receptor (GLP-1R) has a critical role in GLP-1 peptide binding and receptor activation.J Biol Chem. 2012 Feb 3;287(6):3642-58. doi: 10.1074/jbc.M111.309328. Epub 2011 Dec 6. J Biol Chem. 2012. PMID: 22147710 Free PMC article.
References
-
- Al-Sabah S, Donnelly D. The positive charge at Lys-288 of the glucagon-like peptide-1 (GLP-1) receptor is important for binding the N-terminus of peptide agonists. FEBS Lett. 2003b;553:342–346. - PubMed
-
- Baggio LL, Drucker DJ. Biology of incretins: GLP-1 and GIP. Gastroenterology. 2007;132:2131–2157. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

