MZmine 2: modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data
- PMID: 20650010
- PMCID: PMC2918584
- DOI: 10.1186/1471-2105-11-395
MZmine 2: modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data
Abstract
Background: Mass spectrometry (MS) coupled with online separation methods is commonly applied for differential and quantitative profiling of biological samples in metabolomic as well as proteomic research. Such approaches are used for systems biology, functional genomics, and biomarker discovery, among others. An ongoing challenge of these molecular profiling approaches, however, is the development of better data processing methods. Here we introduce a new generation of a popular open-source data processing toolbox, MZmine 2.
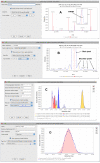
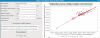
Results: A key concept of the MZmine 2 software design is the strict separation of core functionality and data processing modules, with emphasis on easy usability and support for high-resolution spectra processing. Data processing modules take advantage of embedded visualization tools, allowing for immediate previews of parameter settings. Newly introduced functionality includes the identification of peaks using online databases, MSn data support, improved isotope pattern support, scatter plot visualization, and a new method for peak list alignment based on the random sample consensus (RANSAC) algorithm. The performance of the RANSAC alignment was evaluated using synthetic datasets as well as actual experimental data, and the results were compared to those obtained using other alignment algorithms.
Conclusions: MZmine 2 is freely available under a GNU GPL license and can be obtained from the project website at: http://mzmine.sourceforge.net/. The current version of MZmine 2 is suitable for processing large batches of data and has been applied to both targeted and non-targeted metabolomic analyses.
Figures


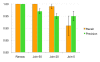
References
-
- Laaksonen R, Katajamaa M, Paiva H, Sysi-Aho M, Saarinen L, Junni P, Lutjohann D, Smet J, Van Coster R, Seppanen-Laakso T, Lehtimäki T, Soini J, Oresic M. A systems biology strategy reveals biological pathways and plasma biomarker candidates for potentially toxic statin-induced changes in muscle. PLoS One. 2006;1:e97. doi: 10.1371/journal.pone.0000097. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

