Comprehensive spectroscopic, steady state, and transient kinetic studies of a representative siderophore-associated flavin monooxygenase
- PMID: 20650894
- PMCID: PMC2945530
- DOI: 10.1074/jbc.M110.157578
Comprehensive spectroscopic, steady state, and transient kinetic studies of a representative siderophore-associated flavin monooxygenase
Abstract
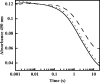
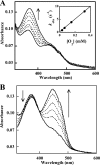
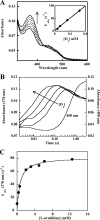
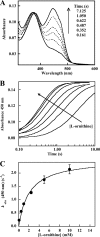
Many siderophores used for the uptake and intracellular storage of essential iron contain hydroxamate chelating groups. Their biosyntheses are typically initiated by hydroxylation of the primary amine side chains of l-ornithine or l-lysine. This reaction is catalyzed by members of a widespread family of FAD-dependent monooxygenases. Here the kinetic mechanism for a representative family member has been extensively characterized by steady state and transient kinetic methods, using heterologously expressed N(5)-l-ornithine monooxygenase from the pathogenic fungus Aspergillus fumigatus. Spectroscopic data and kinetic analyses suggest a model in which a molecule of hydroxylatable substrate serves as an activator for the reaction of the reduced flavin and O(2). The rate acceleration is only ∼5-fold, a mild effect of substrate on formation of the C4a-hydroperoxide that does not influence the overall rate of turnover. The effect is also observed with the bacterial ornithine monooxygenase PvdA. The C4a-hydroperoxide is stabilized in the absence of hydroxylatable substrate by the presence of bound NADP(+) (t(½) = 33 min, 25 °C, pH 8). NADP(+) therefore is a likely regulator of O(2) and substrate reactivity in the siderophore-associated monooxygenases. Aside from the activating effect of the hydroxylatable substrate, the siderophore-associated monooxygenases share a kinetic mechanism with the hepatic microsomal flavin monooxygenases and bacterial Baeyer-Villiger monooxygenases, with which they share only moderate sequence homology and from which they are distinguished by their acute substrate specificity. The remarkable specificity of the N(5)-l-ornithine monooxygenase-catalyzed reaction suggests added means of reaction control beyond those documented in related well characterized flavoenzymes.
Figures

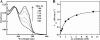

Similar articles
-
Aspergillus fumigatus SidA is a highly specific ornithine hydroxylase with bound flavin cofactor.Biochemistry. 2010 Aug 10;49(31):6777-83. doi: 10.1021/bi100291n. Biochemistry. 2010. PMID: 20614882
-
Arg279 is the key regulator of coenzyme selectivity in the flavin-dependent ornithine monooxygenase SidA.Biochim Biophys Acta. 2014 Apr;1844(4):778-84. doi: 10.1016/j.bbapap.2014.02.005. Epub 2014 Feb 15. Biochim Biophys Acta. 2014. PMID: 24534646
-
Role of Ser-257 in the sliding mechanism of NADP(H) in the reaction catalyzed by the Aspergillus fumigatus flavin-dependent ornithine N5-monooxygenase SidA.J Biol Chem. 2013 Nov 8;288(45):32440-32448. doi: 10.1074/jbc.M113.487181. Epub 2013 Sep 26. J Biol Chem. 2013. PMID: 24072704 Free PMC article.
-
Mechanistic and structural studies of the N-hydroxylating flavoprotein monooxygenases.Bioorg Chem. 2011 Dec;39(5-6):171-7. doi: 10.1016/j.bioorg.2011.07.006. Epub 2011 Aug 5. Bioorg Chem. 2011. PMID: 21871647 Free PMC article. Review.
-
Control of catalysis in flavin-dependent monooxygenases.Arch Biochem Biophys. 2010 Jan 1;493(1):26-36. doi: 10.1016/j.abb.2009.11.028. Epub 2009 Nov 26. Arch Biochem Biophys. 2010. PMID: 19944667 Review.
Cited by
-
Kinetic Mechanism of the Dechlorinating Flavin-dependent Monooxygenase HadA.J Biol Chem. 2017 Mar 24;292(12):4818-4832. doi: 10.1074/jbc.M116.774448. Epub 2017 Feb 3. J Biol Chem. 2017. PMID: 28159841 Free PMC article.
-
Allelic analyses of the Arabidopsis YUC1 locus reveal residues and domains essential for the functions of YUC family of flavin monooxygenases.J Integr Plant Biol. 2011 Jan;53(1):54-62. doi: 10.1111/j.1744-7909.2010.01007.x. Epub 2010 Dec 22. J Integr Plant Biol. 2011. PMID: 21205174 Free PMC article.
-
Beyond the Protein Matrix: Probing Cofactor Variants in a Baeyer-Villiger Oxygenation Reaction.ACS Catal. 2013;3(12):3058-3062. doi: 10.1021/cs400837z. ACS Catal. 2013. PMID: 24443704 Free PMC article.
-
Two structures of an N-hydroxylating flavoprotein monooxygenase: ornithine hydroxylase from Pseudomonas aeruginosa.J Biol Chem. 2011 Sep 9;286(36):31789-98. doi: 10.1074/jbc.M111.265876. Epub 2011 Jul 13. J Biol Chem. 2011. PMID: 21757711 Free PMC article.
-
The reaction kinetics of 3-hydroxybenzoate 6-hydroxylase from Rhodococcus jostii RHA1 provide an understanding of the para-hydroxylation enzyme catalytic cycle.J Biol Chem. 2013 Dec 6;288(49):35210-21. doi: 10.1074/jbc.M113.515205. Epub 2013 Oct 15. J Biol Chem. 2013. PMID: 24129570 Free PMC article.
References
-
- Ong S. T., Ho J. Z., Ho B., Ding J. L. (2006) Immunobiology 211, 295–314 - PubMed
-
- Jurado R. L. (1997) Clin. Infect. Dis. 25, 888–895 - PubMed
-
- Wandersman C., Delepelaire P. (2004) Annu. Rev. Microbiol. 58, 611–647 - PubMed
-
- Andrews S. C., Robinson A. K., Rodríguez-Quiñones F. (2003) FEMS Microbiol. Rev. 27, 215–237 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

