Prophossi: automating expert validation of phosphopeptide-spectrum matches from tandem mass spectrometry
- PMID: 20651112
- PMCID: PMC2922888
- DOI: 10.1093/bioinformatics/btq341
Prophossi: automating expert validation of phosphopeptide-spectrum matches from tandem mass spectrometry
Abstract
Motivation: Complex patterns of protein phosphorylation mediate many cellular processes. Tandem mass spectrometry (MS/MS) is a powerful tool for identifying these post-translational modifications. In high-throughput experiments, mass spectrometry database search engines, such as MASCOT provide a ranked list of peptide identifications based on hundreds of thousands of MS/MS spectra obtained in a mass spectrometry experiment. These search results are not in themselves sufficient for confident assignment of phosphorylation sites as identification of characteristic mass differences requires time-consuming manual assessment of the spectra by an experienced analyst. The time required for manual assessment has previously rendered high-throughput confident assignment of phosphorylation sites challenging.
Results: We have developed a knowledge base of criteria, which replicate expert assessment, allowing more than half of cases to be automatically validated and site assignments verified with a high degree of confidence. This was assessed by comparing automated spectral interpretation with careful manual examination of the assignments for 501 peptides above the 1% false discovery rate (FDR) threshold corresponding to 259 putative phosphorylation sites in 74 proteins of the Trypanosoma brucei proteome. Despite this stringent approach, we are able to validate 80 of the 91 phosphorylation sites (88%) positively identified by manual examination of the spectra used for the MASCOT searches with a FDR < 15%.
Conclusions: High-throughput computational analysis can provide a viable second stage validation of primary mass spectrometry database search results. Such validation gives rapid access to a systems level overview of protein phosphorylation in the experiment under investigation.
Availability: A GPL licensed software implementation in Perl for analysis and spectrum annotation is available in the supplementary material and a web server can be assessed online at http://www.compbio.dundee.ac.uk/prophossi.
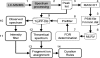
Figures



Similar articles
-
SimPhospho: a software tool enabling confident phosphosite assignment.Bioinformatics. 2018 Aug 1;34(15):2690-2692. doi: 10.1093/bioinformatics/bty151. Bioinformatics. 2018. PMID: 29596608 Free PMC article.
-
Automatic validation of phosphopeptide identifications from tandem mass spectra.Anal Chem. 2007 Feb 15;79(4):1301-10. doi: 10.1021/ac061334v. Anal Chem. 2007. PMID: 17297928 Free PMC article.
-
Integrated data management and validation platform for phosphorylated tandem mass spectrometry data.Proteomics. 2010 Oct;10(19):3515-24. doi: 10.1002/pmic.200900727. Proteomics. 2010. PMID: 20827731 Free PMC article.
-
The spectral networks paradigm in high throughput mass spectrometry.Mol Biosyst. 2012 Oct;8(10):2535-44. doi: 10.1039/c2mb25085c. Mol Biosyst. 2012. PMID: 22610447 Free PMC article. Review.
-
Open source libraries and frameworks for mass spectrometry based proteomics: a developer's perspective.Biochim Biophys Acta. 2014 Jan;1844(1 Pt A):63-76. doi: 10.1016/j.bbapap.2013.02.032. Epub 2013 Mar 1. Biochim Biophys Acta. 2014. PMID: 23467006 Free PMC article. Review.
Cited by
-
On the Extent of Tyrosine Phosphorylation in Chloroplasts.Plant Physiol. 2015 Oct;169(2):996-1000. doi: 10.1104/pp.15.00921. Epub 2015 Aug 4. Plant Physiol. 2015. PMID: 26243617 Free PMC article.
-
Selective chemoprecipitation to enrich nitropeptides from complex proteomes for mass-spectrometric analysis.Nat Protoc. 2014 Apr;9(4):882-95. doi: 10.1038/nprot.2014.052. Epub 2014 Mar 20. Nat Protoc. 2014. PMID: 24651500 Free PMC article.
-
PhosphoHunter: An Efficient Software Tool for Phosphopeptide Identification.Adv Bioinformatics. 2015;2015:382869. doi: 10.1155/2015/382869. Epub 2015 Jan 12. Adv Bioinformatics. 2015. PMID: 25653679 Free PMC article.
-
Quantitative phosphoproteomics unravels biased phosphorylation of serotonin 2A receptor at Ser280 by hallucinogenic versus nonhallucinogenic agonists.Mol Cell Proteomics. 2014 May;13(5):1273-85. doi: 10.1074/mcp.M113.036558. Epub 2014 Mar 17. Mol Cell Proteomics. 2014. PMID: 24637012 Free PMC article.
-
A Naturally Occurring Urinary Collagen Type I Alpha 1-Derived Peptide Inhibits Collagen Type I-Induced Endothelial Cell Migration at Physiological Concentrations.Int J Mol Sci. 2025 Aug 2;26(15):7480. doi: 10.3390/ijms26157480. Int J Mol Sci. 2025. PMID: 40806611 Free PMC article.
References
-
- Andersson L, Porath J. Isolation of phosphoproteins by immobilized metal (Fe3+) affinity chromatography. Anal. Biochem. 1986;154:250–254. - PubMed
-
- Beausoleil SA, et al. A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nat. Biotechnol. 2006;24:1285–1292. - PubMed
-
- Breci LA, et al. Cleavage N-terminal to proline: analysis of a database of peptide tandem mass spectra. Anal. Chem. 2003;75:1963–1971. - PubMed
-
- Cohen P. The regulation of protein function by multisite phosphorylation—a 25 year update. Trends Biochem. Sci. 2000;25:596–601. - PubMed
-
- Cox J, Mann M. Is proteomics the new genomics? Cell. 2007;130:395–398. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

