Voltage-gated Na+ channel SCN5A is a key regulator of a gene transcriptional network that controls colon cancer invasion
- PMID: 20651255
- PMCID: PMC2936697
- DOI: 10.1158/0008-5472.CAN-10-1169
Voltage-gated Na+ channel SCN5A is a key regulator of a gene transcriptional network that controls colon cancer invasion
Abstract
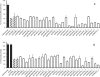
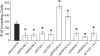
Voltage-gated Na(+) channels (VGSC) have been implicated in the metastatic potential of human breast, prostate, and lung cancer cells. Specifically, the SCN5A gene encoding the VGSC isotype Na(v)1.5 has been defined as a key driver of human cancer cell invasion. In this study, we examined the expression and function of VGSCs in a panel of colon cancer cell lines by electrophysiologic recordings. Na(+) channel activity and invasive potential were inhibited pharmacologically by tetrodotoxin or genetically by small interfering RNAs (siRNA) specifically targeting SCN5A. Clinical relevance was established by immunohistochemistry of patient biopsies, with strong Na(v)1.5 protein staining found in colon cancer specimens but little to no staining in matched-paired normal colon tissues. We explored the mechanism of VGSC-mediated invasive potential on the basis of reported links between VGSC activity and gene expression in excitable cells. Probabilistic modeling of loss-of-function screens and microarray data established an unequivocal role of VGSC SCN5A as a high level regulator of a colon cancer invasion network, involving genes that encompass Wnt signaling, cell migration, ectoderm development, response to biotic stimulus, steroid metabolic process, and cell cycle control. siRNA-mediated knockdown of predicted downstream network components caused a loss of invasive behavior, demonstrating network connectivity and its function in driving colon cancer invasion.
Figures




References
-
- Kunzelmann K. Ion channels and cancer. J Membr Biol. 2005;205(3):159–73. - PubMed
-
- Schuller HM. Is cancer triggered by altered signalling of nicotinic acetylcholine receptors? Nature reviews. 2009;9(3):195–205. - PubMed
-
- Onkal R, Djamgoz MB. Molecular pharmacology of voltage-gated sodium channel expression in metastatic disease: clinical potential of neonatal Nav1.5 in breast cancer. European journal of pharmacology. 2009;625(1–3):206–19. - PubMed
-
- Roger S, Potier M, Vandier C, Besson P, Le Guennec JY. Voltage-gated sodium channels: new targets in cancer therapy? Curr Pharm Des. 2006;12(28):3681–95. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

