Cryptic variation between species and the basis of hybrid performance
- PMID: 20652019
- PMCID: PMC2907293
- DOI: 10.1371/journal.pbio.1000429
Cryptic variation between species and the basis of hybrid performance
Abstract
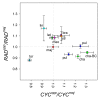
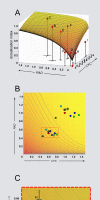
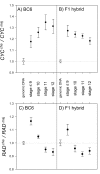
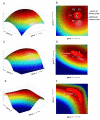
Crosses between closely related species give two contrasting results. One result is that species hybrids may be inferior to their parents, for example, being less fertile [1]. The other is that F1 hybrids may display superior performance (heterosis), for example with increased vigour [2]. Although various hypotheses have been proposed to account for these two aspects of hybridisation, their biological basis is still poorly understood [3]. To gain further insights into this issue, we analysed the role that variation in gene expression may play. We took a conserved trait, flower asymmetry in Antirrhinum, and determined the extent to which the underlying regulatory genes varied in expression among closely related species. We show that expression of both genes analysed, CYC and RAD, varies significantly between species because of cis-acting differences. By making a quantitative genotype-phenotype map, using a range of mutant alleles, we demonstrate that the species lie on a plateau in gene expression-morphology space, so that the variation has no detectable phenotypic effect. However, phenotypic differences can be revealed by shifting genotypes off the plateau through genetic crosses. Our results can be readily explained if genomes are free to evolve within an effectively neutral zone in gene expression space. The consequences of this drift will be negligible for individual loci, but when multiple loci across the genome are considered, we show that the variation may have significant effects on phenotype and fitness, causing a significant drift load. By considering these consequences for various gene-expression-fitness landscapes, we conclude that F1 hybrids might be expected to show increased performance with regard to conserved traits, such as basic physiology, but reduced performance with regard to others. Thus, our study provides a new way of explaining how various aspects of hybrid performance may arise through natural variation in gene activity.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



Comment in
-
Population genetics: Breaking down hybrids.Nat Rev Genet. 2010 Sep;11(9):594. doi: 10.1038/nrg2854. Nat Rev Genet. 2010. PMID: 20799390 No abstract available.
References
-
- Orr H. A, Presgraves D. C. Speciation by postzygotic isolation: forces, genes and molecules. Bioessays. 2000;22:1085–1094. - PubMed
-
- Burke J. M, Arnold M. L. Genetics and the Fitness of Hybrids. Annu Rev Genet. 2001;35:31–52. - PubMed
-
- Bateson W. Heredity and variation in modern lights. In: Seward A. C, editor. Darwin and modern science. Cambridge: Cambridge University Press; 1909.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

