ADP-ribosylation of arginine
- PMID: 20652610
- PMCID: PMC3102197
- DOI: 10.1007/s00726-010-0676-2
ADP-ribosylation of arginine
Abstract
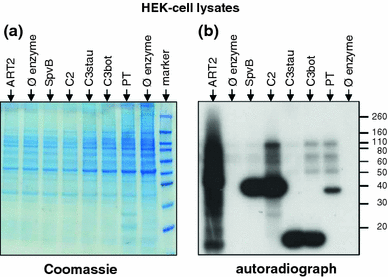
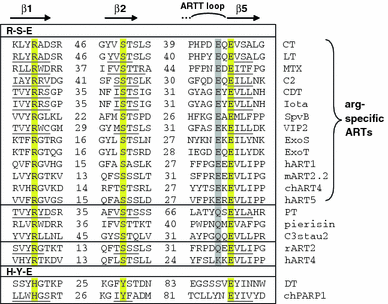
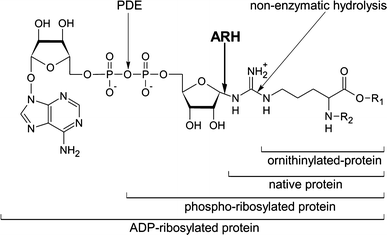
Arginine adenosine-5'-diphosphoribosylation (ADP-ribosylation) is an enzyme-catalyzed, potentially reversible posttranslational modification, in which the ADP-ribose moiety is transferred from NAD(+) to the guanidino moiety of arginine. At 540 Da, ADP-ribose has the size of approximately five amino acid residues. In contrast to arginine, which, at neutral pH, is positively charged, ADP-ribose carries two negatively charged phosphate moieties. Arginine ADP-ribosylation, thus, causes a notable change in size and chemical property at the ADP-ribosylation site of the target protein. Often, this causes steric interference of the interaction of the target protein with binding partners, e.g. toxin-catalyzed ADP-ribosylation of actin at R177 sterically blocks actin polymerization. In case of the nucleotide-gated P2X7 ion channel, ADP-ribosylation at R125 in the vicinity of the ligand-binding site causes channel gating. Arginine-specific ADP-ribosyltransferases (ARTs) carry a characteristic R-S-EXE motif that distinguishes these enzymes from structurally related enzymes which catalyze ADP-ribosylation of other amino acid side chains, DNA, or small molecules. Arginine-specific ADP-ribosylation can be inhibited by small molecule arginine analogues such as agmatine or meta-iodobenzylguanidine (MIBG), which themselves can serve as targets for arginine-specific ARTs. ADP-ribosylarginine specific hydrolases (ARHs) can restore target protein function by hydrolytic removal of the entire ADP-ribose moiety. In some cases, ADP-ribosylarginine is processed into secondary posttranslational modifications, e.g. phosphoribosylarginine or ornithine. This review summarizes current knowledge on arginine-specific ADP-ribosylation, focussing on the methods available for its detection, its biological consequences, and the enzymes responsible for this modification and its reversal, and discusses future perspectives for research in this field.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

