The ESCRT complexes
- PMID: 20653365
- PMCID: PMC2988974
- DOI: 10.3109/10409238.2010.502516
The ESCRT complexes
Abstract
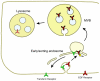
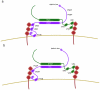
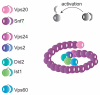
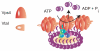
The ESCRT machinery consists of the peripheral membrane protein complexes ESCRT-0, -I, -II, -III, and Vps4-Vta1, and the ALIX homodimer. The ESCRT system is required for degradation of unneeded or dangerous plasma membrane proteins; biogenesis of the lysosome and the yeast vacuole; the budding of most membrane enveloped viruses; the membrane abscission step in cytokinesis; macroautophagy; and several other processes. From their initial discovery in 2001-2002, the literature on ESCRTs has grown exponentially. This review will describe the structure and function of the six complexes noted above and summarize current knowledge of their mechanistic roles in cellular pathways and in disease.
Figures








References
-
- Agromayor M, Martin-Serrano J. Interaction of AMSH with ESCRT-III and deubiquitination of endosomal cargo. J Biol Chem. 2006;281:23083–23091. - PubMed
-
- Alam SL, Langelier C, Whitby FG, Koirala S, Robinson H, Hill CP, Sundquist WI. Structural basis for ubiquitin recognition by the human ESCRT-II EAP45 GLUE domain. Nat Struct Mol Biol. 2006;13:1029–1030. - PubMed
-
- Alam SL, Sundquist WI. Structural biology - ESCRT service. Nature. 2007;447:921–922. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
