Application of machine learning methods to histone methylation ChIP-Seq data reveals H4R3me2 globally represses gene expression
- PMID: 20653935
- PMCID: PMC2928206
- DOI: 10.1186/1471-2105-11-396
Application of machine learning methods to histone methylation ChIP-Seq data reveals H4R3me2 globally represses gene expression
Abstract
Background: In the last decade, biochemical studies have revealed that epigenetic modifications including histone modifications, histone variants and DNA methylation form a complex network that regulate the state of chromatin and processes that depend on it including transcription and DNA replication. Currently, a large number of these epigenetic modifications are being mapped in a variety of cell lines at different stages of development using high throughput sequencing by members of the ENCODE consortium, the NIH Roadmap Epigenomics Program and the Human Epigenome Project. An extremely promising and underexplored area of research is the application of machine learning methods, which are designed to construct predictive network models, to these large-scale epigenomic data sets.
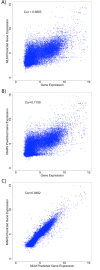
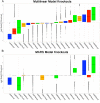
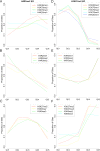
Results: Using a ChIP-Seq data set of 20 histone lysine and arginine methylations and histone variant H2A.Z in human CD4+ T-cells, we built predictive models of gene expression as a function of histone modification/variant levels using Multilinear (ML) Regression and Multivariate Adaptive Regression Splines (MARS). Along with extensive crosstalk among the 20 histone methylations, we found H4R3me2 was the most and second most globally repressive histone methylation among the 20 studied in the ML and MARS models, respectively. In support of our finding, a number of experimental studies show that PRMT5-catalyzed symmetric dimethylation of H4R3 is associated with repression of gene expression. This includes a recent study, which demonstrated that H4R3me2 is required for DNMT3A-mediated DNA methylation--a known global repressor of gene expression.
Conclusion: In stark contrast to univariate analysis of the relationship between H4R3me2 and gene expression levels, our study showed that the regulatory role of some modifications like H4R3me2 is masked by confounding variables, but can be elucidated by multivariate/systems-level approaches.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

