Tracing phylogenomic events leading to diversity of Haemophilus influenzae and the emergence of Brazilian Purpuric Fever (BPF)-associated clones
- PMID: 20654709
- PMCID: PMC2967034
- DOI: 10.1016/j.ygeno.2010.07.005
Tracing phylogenomic events leading to diversity of Haemophilus influenzae and the emergence of Brazilian Purpuric Fever (BPF)-associated clones
Abstract
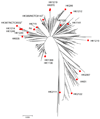
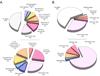
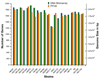
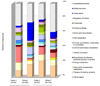
Here we report the use of a multi-genome DNA microarray to elucidate the genomic events associated with the emergence of the clonal variants of Haemophilus influenzae biogroup aegyptius causing Brazilian Purpuric Fever (BPF), an important pediatric disease with a high mortality rate. We performed directed genome sequencing of strain HK1212 unique loci to construct a species DNA microarray. Comparative genome hybridization using this microarray enabled us to determine and compare gene complements, and infer reliable phylogenomic relationships among members of the species. The higher genomic variability observed in the genomes of BPF-related strains (clones) and their close relatives may be characterized by significant gene flux related to a subset of functional role categories. We found that the acquisition of a large number of virulence determinants featuring numerous cell membrane proteins coupled to the loss of genes involved in transport, central biosynthetic pathways and in particular, energy production pathways to be characteristics of the BPF genomic variants.
Copyright © 2010 Elsevier Inc. All rights reserved.
Figures



Similar articles
-
Identification and characterization of genomic loci unique to the Brazilian purpuric fever clonal group of H. influenzae biogroup aegyptius: functionality explored using meningococcal homology.Mol Microbiol. 2003 Feb;47(4):1101-11. doi: 10.1046/j.1365-2958.2003.03359.x. Mol Microbiol. 2003. PMID: 12581362
-
Biochemical, genetic, and epidemiologic characterization of Haemophilus influenzae biogroup aegyptius (Haemophilus aegyptius) strains associated with Brazilian purpuric fever.J Clin Microbiol. 1988 Aug;26(8):1524-34. doi: 10.1128/jcm.26.8.1524-1534.1988. J Clin Microbiol. 1988. PMID: 3262623 Free PMC article.
-
Genomic analysis of the F3031 Brazilian purpuric fever clone of Haemophilus influenzae biogroup aegyptius by PCR-based subtractive hybridization.Infect Immun. 2002 May;70(5):2694-9. doi: 10.1128/IAI.70.5.2694-2699.2002. Infect Immun. 2002. PMID: 11953414 Free PMC article.
-
Emergence and disappearance of a virulent clone of Haemophilus influenzae biogroup aegyptius, cause of Brazilian purpuric fever.Clin Microbiol Rev. 2008 Oct;21(4):594-605. doi: 10.1128/CMR.00020-08. Clin Microbiol Rev. 2008. PMID: 18854482 Free PMC article. Review.
-
Brazilian purpuric fever identified in a new region of Brazil. The Brazilian Purpuric Fever Study Group.J Infect Dis. 1992 Jun;165 Suppl 1:S16-9. doi: 10.1093/infdis/165-supplement_1-s16. J Infect Dis. 1992. PMID: 1588153 Review.
Cited by
-
Investigating the genome diversity of B. cereus and evolutionary aspects of B. anthracis emergence.Genomics. 2011 Jul;98(1):26-39. doi: 10.1016/j.ygeno.2011.03.008. Epub 2011 Apr 5. Genomics. 2011. PMID: 21447378 Free PMC article.
-
Comparative genomic analysis reveals distinct genotypic features of the emerging pathogen Haemophilus influenzae type f.BMC Genomics. 2014 Jan 18;15(1):38. doi: 10.1186/1471-2164-15-38. BMC Genomics. 2014. PMID: 24438474 Free PMC article.
-
Emerging infectious diseases: threats to human health and global stability.PLoS Pathog. 2013;9(7):e1003467. doi: 10.1371/journal.ppat.1003467. Epub 2013 Jul 4. PLoS Pathog. 2013. PMID: 23853589 Free PMC article. No abstract available.
-
Classification, identification, and clinical significance of Haemophilus and Aggregatibacter species with host specificity for humans.Clin Microbiol Rev. 2014 Apr;27(2):214-40. doi: 10.1128/CMR.00103-13. Clin Microbiol Rev. 2014. PMID: 24696434 Free PMC article. Review.
References
-
- Kilian M. Haemophilus. In: Murray PR, Baron EJ, Jorgensen JH, Landry ML, Pfaller MA, editors. Manual of Clinial Microbiology. 9th ed. Washinton D.C.: American Society of Microbiology; 2007. pp. 636–648.
-
- Kroll JS, Loynds B, Brophy LN, Moxon ER. The bex locus in encapsulated Haemophilus influenzae: a chromosomal region involved in capsule polysaccharide export. Mol Microbiol. 1990;4:1853–1862. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

