The MRN complex in double-strand break repair and telomere maintenance
- PMID: 20655309
- PMCID: PMC2946096
- DOI: 10.1016/j.febslet.2010.07.029
The MRN complex in double-strand break repair and telomere maintenance
Abstract
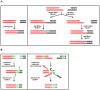
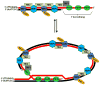
Genomes are subject to constant threat by damaging agents that generate DNA double-strand breaks (DSBs). The ends of linear chromosomes need to be protected from DNA damage recognition and end-joining, and this is achieved through protein-DNA complexes known as telomeres. The Mre11-Rad50-Nbs1 (MRN) complex plays important roles in detection and signaling of DSBs, as well as the repair pathways of homologous recombination (HR) and non-homologous end-joining (NHEJ). In addition, MRN associates with telomeres and contributes to their maintenance. Here, we provide an overview of MRN functions at DSBs, and examine its roles in telomere maintenance and dysfunction.
Copyright 2010 Federation of European Biochemical Societies. Published by Elsevier B.V. All rights reserved.
Figures


References
-
- Shrivastav M, De Haro LP, Nickoloff JA. Regulation of DNA double-strand break repair pathway choice. Cell Res. 2008;18:134–47. - PubMed
-
- Rass E, Grabarz A, Plo I, Gautier J, Bertrand P, Lopez BS. Role of Mre11 in chromosomal nonhomologous end joining in mammalian cells. Nat Struct Mol Biol. 2009;16:819–24. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

