Differential regulation of JAMM domain deubiquitinating enzyme activity within the RAP80 complex
- PMID: 20656689
- PMCID: PMC2945588
- DOI: 10.1074/jbc.M110.135319
Differential regulation of JAMM domain deubiquitinating enzyme activity within the RAP80 complex
Abstract
BRCC36 is a JAMM (JAB1/MPN/Mov34 metalloenzyme) domain, lysine 63-ubiquitin (K63-Ub)-specific deubiquitinating enzyme (DUB) and a member of two protein complexes: the DNA damage-responsive BRCA1-RAP80 complex, and the cytoplasmic BRCC36 isopeptidase complex (BRISC). The presence of several identical constituents in both complexes suggests common regulatory mechanisms and potential competition between K63-Ub-related signaling in cytoplasmic and nuclear compartments. Surprisingly, we discover that BRCC36 DUB activity requires different interactions within the context of each complex. Abraxas and BRCC45 were essential for BRCC36 DUB activity within the RAP80 complex, whereas KIAA0157/Abro was the only interaction required for DUB activity within the BRISC. Poh1 also required protein interactions for activity, suggesting a common regulatory mechanism for JAMM domain DUBs. Finally, BRISC deficiency enhanced formation of the BRCA1-RAP80 complex in vivo, increasing BRCA1 levels at DNA double strand breaks. These findings reveal that JAMM domain DUB activity and K63-Ub levels are regulated by multiple mechanisms within the cell.
Figures

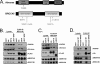





References
-
- Aguilera A., Gómez-González B. (2008) Nat. Rev. Genet. 9, 204–217 - PubMed
-
- Zhou B. B., Elledge S. J. (2000) Nature 408, 433–439 - PubMed
-
- Bartek J., Lukas J. (2007) Curr. Opin. Cell Biol. 19, 238–245 - PubMed
-
- Peng J., Schwartz D., Elias J. E., Thoreen C. C., Cheng D., Marsischky G., Roelofs J., Finley D., Gygi S. P. (2003) Nat. Biotechnol. 21, 921–926 - PubMed
-
- Pickart C. M., Cohen R. E. (2004) Nat. Rev. Mol. Cell Biol. 5, 177–187 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

