A group II intron encodes a functional LAGLIDADG homing endonuclease and self-splices under moderate temperature and ionic conditions
- PMID: 20656798
- PMCID: PMC2924541
- DOI: 10.1261/rna.2184010
A group II intron encodes a functional LAGLIDADG homing endonuclease and self-splices under moderate temperature and ionic conditions
Abstract
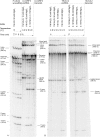
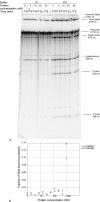
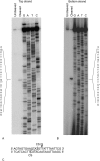
A group II intron encoding a protein belonging to the LAGLIDADG family of homing endonucleases was identified in the mitochondrial rns gene of the filamentous fungus Leptographium truncatum, and the catalytic activities of both the intron and its encoded protein were characterized. A model of the RNA secondary structure indicates that the intron is a member of the IIB1 subclass and the open reading frame is inserted in ribozyme domain III. In vitro assays carried out with two versions of the intron, one in which the open reading frame was removed and the other in which it was present, demonstrate that both versions of the intron readily self-splice at 37 degrees C and at a concentration of MgCl(2) as low as 6 mM. The open reading frame encodes a functional LAGLIDADG homing endonuclease that cleaves 2 (top strand) and 6 (bottom strand) nucleotides (nt) upstream of the intron insertion site, generating 4 nt 3' OH overhangs. In vitro splicing assays carried out in the absence and presence of the intron-encoded protein indicate that the protein does not enhance intron splicing, and RNA-binding assays show that the protein does not appear to bind to the intron RNA precursor transcript. These findings raise intriguing questions concerning the functional and evolutionary relationships of the two components of this unique composite element.
Figures



Similar articles
-
I-OmiI and I-OmiII: two intron-encoded homing endonucleases within the Ophiostoma minus rns gene.Fungal Biol. 2014 Aug;118(8):721-31. doi: 10.1016/j.funbio.2014.05.002. Epub 2014 May 23. Fungal Biol. 2014. PMID: 25110134
-
Evolutionary dynamics of the mS952 intron: a novel mitochondrial group II intron encoding a LAGLIDADG homing endonuclease gene.J Mol Evol. 2011 Jun;72(5-6):433-49. doi: 10.1007/s00239-011-9442-7. Epub 2011 Apr 10. J Mol Evol. 2011. PMID: 21479820
-
Expression of the Naegleria intron endonuclease is dependent on a functional group I self-cleaving ribozyme.RNA. 2000 Apr;6(4):616-27. doi: 10.1017/s1355838200992203. RNA. 2000. PMID: 10786852 Free PMC article.
-
Group II introns: structure and catalytic versatility of large natural ribozymes.Crit Rev Biochem Mol Biol. 2003;38(3):249-303. doi: 10.1080/713609236. Crit Rev Biochem Mol Biol. 2003. PMID: 12870716 Review.
-
Homing endonuclease structure and function.Q Rev Biophys. 2005 Feb;38(1):49-95. doi: 10.1017/S0033583505004063. Epub 2005 Dec 9. Q Rev Biophys. 2005. PMID: 16336743 Review.
Cited by
-
A new RNA-DNA interaction required for integration of group II intron retrotransposons into DNA targets.Nucleic Acids Res. 2021 Dec 2;49(21):12394-12410. doi: 10.1093/nar/gkab1031. Nucleic Acids Res. 2021. PMID: 34791436 Free PMC article.
-
Protein-free catalysis of DNA hydrolysis and self-integration by a ribozyme.Nucleic Acids Res. 2025 Jan 11;53(2):gkae1224. doi: 10.1093/nar/gkae1224. Nucleic Acids Res. 2025. PMID: 39698822 Free PMC article.
-
Prevalence of Group II Introns in Phage Genomes.bioRxiv [Preprint]. 2025 May 23:2025.05.22.655115. doi: 10.1101/2025.05.22.655115. bioRxiv. 2025. Update in: Nucleic Acids Res. 2025 Aug 11;53(15):gkaf761. doi: 10.1093/nar/gkaf761. PMID: 40475605 Free PMC article. Updated. Preprint.
-
Evidence of Extensive Intraspecific Noncoding Reshuffling in a 169-kb Mitochondrial Genome of a Basidiomycetous Fungus.Genome Biol Evol. 2019 Oct 1;11(10):2774-2788. doi: 10.1093/gbe/evz181. Genome Biol Evol. 2019. PMID: 31418013 Free PMC article.
-
Recurrent insertion of 5'-terminal nucleotides and loss of the branchpoint motif in lineages of group II introns inserted in mitochondrial preribosomal RNAs.RNA. 2011 Jul;17(7):1321-35. doi: 10.1261/rna.2655911. Epub 2011 May 25. RNA. 2011. PMID: 21613530 Free PMC article.
References
-
- Bae H, Kim KP, Song JM, Kim JH, Yang JS, Kwon ST 2009. Characterization of intein homing endonuclease encoded in the DNA polymerase gene of Thermococcus marinus. FEMS Microbiol Lett 297: 180–188 - PubMed
-
- Bassi GS, Weeks KM 2003. Kinetic and thermodynamic framework for assembly of the six-component bI3 group I ribonucleoprotein catalyst. Biochemistry 42: 9980–9988 - PubMed
-
- Belfort M 2003. Two for the price of one: a bifunctional intron-encoded DNA endonuclease-RNA maturase. Genes Dev 17: 2860–2863 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
