Characterization of the PduS cobalamin reductase of Salmonella enterica and its role in the Pdu microcompartment
- PMID: 20656910
- PMCID: PMC2944522
- DOI: 10.1128/JB.00575-10
Characterization of the PduS cobalamin reductase of Salmonella enterica and its role in the Pdu microcompartment
Abstract
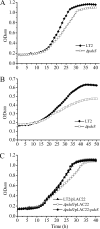
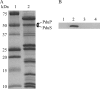
Salmonella enterica degrades 1,2-propanediol (1,2-PD) in a coenzyme B12 (adenosylcobalamin, AdoCbl)-dependent fashion. Salmonella obtains AdoCbl by assimilation of complex precursors, such as vitamin B12 and hydroxocobalamin. Assimilation of these compounds requires reduction of their central cobalt atom from Co3+ to Co2+ to Co+, followed by adenosylation to AdoCbl. In this work, the His6-tagged PduS cobalamin reductase from S. enterica was produced at high levels in Escherichia coli, purified, and characterized. The anaerobically purified enzyme reduced cob(III)alamin to cob(II)alamin at a rate of 42.3±3.2 μmol min(-1) mg(-1), and it reduced cob(II)alamin to cob(I)alamin at a rate of 54.5±4.2 nmol min(-1) mg(-1) protein. The apparent Km values of PduS-His6 were 10.1±0.7 μM for NADH and 67.5±8.2 μM for hydroxocobalamin in cob(III)alamin reduction. The apparent Km values for cob(II)alamin reduction were 27.5±2.4 μM with NADH as the substrate and 72.4±9.5 μM with cob(II)alamin as the substrate. High-performance liquid chromatography (HPLC) and mass spectrometry (MS) indicated that each monomer of PduS contained one molecule of noncovalently bound flavin mononucleotide (FMN). Genetic studies showed that a pduS deletion decreased the growth rate of Salmonella on 1,2-PD, supporting a role in cobalamin reduction in vivo. Further studies demonstrated that the PduS protein is a component of the Pdu microcompartments (MCPs) used for 1,2-PD degradation and that it interacts with the PduO adenosyltransferase, which catalyzes the terminal step of AdoCbl synthesis. These studies further characterize PduS, an unusual MCP-associated cobalamin reductase, and, in conjunction with prior results, indicate that the Pdu MCP encapsulates a complete cobalamin assimilation system.
Figures



References
-
- Banerjee, R. 2006. B12 trafficking in mammals: a case for coenzyme escort service. ACS Chem. Biol. 1:149-159. - PubMed
-
- Banerjee, R. (ed.). 1999. Chemistry and Biochemistry of B12. John Wiley and Sons, New York, NY.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

