Quantum chemical calculations on the interaction between flavonol and functional monomers (methacrylic acid and 4-vinylpyridine) in molecularly imprinted polymers
- PMID: 20657423
- PMCID: PMC6264167
- DOI: 10.3390/molecules15064017
Quantum chemical calculations on the interaction between flavonol and functional monomers (methacrylic acid and 4-vinylpyridine) in molecularly imprinted polymers
Abstract
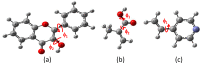
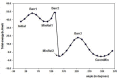
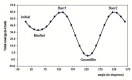
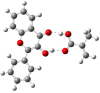
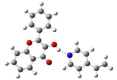
Quantum chemical calculations were performed to characterize the interaction of the flavonol molecule (FL) with methacrylic acid (MAA) and 4-vinylpyridine (4VPy) in the formation of imprinted polymers. The polarizable continuum model (PCM) was used to gain insight on the type of interaction between the reactant molecules under vacuum conditions and in the presence of different solvents. The effect of solvent on the pre-polymerization complex formation was evaluated through the stability energy, in which chloroform behaves as the best solvent for the synthesis of the imprinted polymers since it facilitates the reaction by lowering its degree of stabilization. The reactivity was analyzed in terms of the electrostatic surface potential (ESP) and Mulliken charge. By means of these results, it has been possible to determine two potential recognition sites for the interaction of the MAA monomer and one for the 4VPy in relation to the strength of interaction with FL. In this concern, the interaction of the system FL-MAA is stronger than FL-4VPy.
Figures



Similar articles
-
Study on Dicyandiamide-Imprinted Polymers with Computer-Aided Design.Int J Mol Sci. 2016 Oct 26;17(11):1750. doi: 10.3390/ijms17111750. Int J Mol Sci. 2016. PMID: 27792186 Free PMC article.
-
Synthesis of molecularly imprinted polymers for amino acid derivates by using different functional monomers.Int J Mol Sci. 2011;12(3):1735-43. doi: 10.3390/ijms12031735. Epub 2011 Mar 7. Int J Mol Sci. 2011. PMID: 21673919 Free PMC article.
-
Theoretical and experimental research on the self-assembled system of molecularly imprinted polymers formed by salbutamol and methacrylic acid.J Sep Sci. 2015 Mar;38(6):1065-71. doi: 10.1002/jssc.201401309. Epub 2015 Feb 4. J Sep Sci. 2015. PMID: 25580930
-
Theoretical Insight into the Interaction between Chloramphenicol and Functional Monomer (Methacrylic Acid) in Molecularly Imprinted Polymers.Int J Mol Sci. 2020 Jun 10;21(11):4139. doi: 10.3390/ijms21114139. Int J Mol Sci. 2020. PMID: 32532004 Free PMC article.
-
A brief review of coarse-grained and other computational studies of molecularly imprinted polymers.J Mol Recognit. 2011 Nov-Dec;24(6):883-91. doi: 10.1002/jmr.1135. J Mol Recognit. 2011. PMID: 22038796 Review.
Cited by
-
A novel and highly regioselective synthesis of new carbamoylcarboxylic acids from dianhydrides.ScientificWorldJournal. 2014 Jan 6;2014:725981. doi: 10.1155/2014/725981. eCollection 2014. ScientificWorldJournal. 2014. PMID: 24511299 Free PMC article.
-
Computational Investigation of the Monomer Ratio and Solvent Environment for the Complex Formed between Sulfamethoxazole and Functional Monomer Methacrylic Acid.ACS Omega. 2022 May 10;7(20):17175-17184. doi: 10.1021/acsomega.2c00862. eCollection 2022 May 24. ACS Omega. 2022. PMID: 35647456 Free PMC article.
-
Screening of functional monomers and solvents for the molecular imprinting of paclitaxel separation: a theoretical study.J Mol Model. 2020 Jan 11;26(2):26. doi: 10.1007/s00894-019-4277-z. J Mol Model. 2020. PMID: 31927620
References
-
- Kirsch N., Alexander C., Lübke M., Whitcombe M.J., Vulfson E.N. Enhancement of selectivity of imprinted polymers via post-imprinting modification of recognition sites. Polymer. 2000;41:5583–5590. doi: 10.1016/S0032-3861(99)00782-X. - DOI
-
- Andersson H.S., Koch-Schmidt A.C., Ohlson S., Mosbach K. Study of the nature of recognition in molecularly imprinted polymers. J. Mol. Recognit. 1996;9:675–682. - PubMed
-
- Katz A., Davis M.E. Investigations into the mechanism of molecular recognition with imprinted polymers. Macromolecules. 1999;32:4113–4121. doi: 10.1021/ma981445z. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

