4D-QSAR: perspectives in drug design
- PMID: 20657478
- PMCID: PMC6263259
- DOI: 10.3390/molecules15053281
4D-QSAR: perspectives in drug design
Abstract
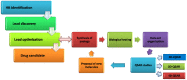
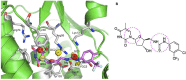
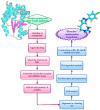
Drug design is a process driven by innovation and technological breakthroughs involving a combination of advanced experimental and computational methods. A broad variety of medicinal chemistry approaches can be used for the identification of hits, generation of leads, as well as to accelerate the optimization of leads into drug candidates. The quantitative structure-activity relationship (QSAR) formalisms are among the most important strategies that can be applied for the successful design new molecules. This review provides a comprehensive review on the evolution and current status of 4D-QSAR, highlighting present challenges and new opportunities in drug design.
Figures

Similar articles
-
Fragment-based QSAR: perspectives in drug design.Mol Divers. 2009 Aug;13(3):277-85. doi: 10.1007/s11030-009-9112-5. Epub 2009 Jan 31. Mol Divers. 2009. PMID: 19184499 Review.
-
Structure-based drug design strategies in medicinal chemistry.Curr Top Med Chem. 2009;9(9):771-90. doi: 10.2174/156802609789207127. Curr Top Med Chem. 2009. PMID: 19754394 Review.
-
Editorial: Current status and perspective on drug targets in tubercle bacilli and drug design of antituberculous agents based on structure-activity relationship.Curr Pharm Des. 2014;20(27):4305-6. doi: 10.2174/1381612819666131118203915. Curr Pharm Des. 2014. PMID: 24245755
-
Applications of Quantitative Structure-Activity Relationships (QSAR) based Virtual Screening in Drug Design: A Review.Mini Rev Med Chem. 2020;20(14):1375-1388. doi: 10.2174/1389557520666200429102334. Mini Rev Med Chem. 2020. PMID: 32348219 Review.
-
Virtual screening and its integration with modern drug design technologies.Curr Med Chem. 2008;15(1):37-46. doi: 10.2174/092986708783330683. Curr Med Chem. 2008. PMID: 18220761 Review.
Cited by
-
QSAR models for removal rates of organic pollutants adsorbed by in situ formed manganese dioxide under acid condition.Environ Sci Pollut Res Int. 2016 Feb;23(4):3609-20. doi: 10.1007/s11356-015-5569-1. Epub 2015 Oct 21. Environ Sci Pollut Res Int. 2016. PMID: 26490942
-
Dihydropyrazole Derivatives Act as Potent α-Amylase Inhibitors and Free Radical Scavengers: Synthesis, Bioactivity Evaluation, Structure-Activity Relationship, ADMET, and Molecular Docking Studies.ACS Omega. 2023 Jun 2;8(23):20412-20422. doi: 10.1021/acsomega.3c00529. eCollection 2023 Jun 13. ACS Omega. 2023. PMID: 37332823 Free PMC article.
-
Comprehensive strategies of machine-learning-based quantitative structure-activity relationship models.iScience. 2021 Aug 28;24(9):103052. doi: 10.1016/j.isci.2021.103052. eCollection 2021 Sep 24. iScience. 2021. PMID: 34553136 Free PMC article. Review.
-
Prevention of Deficit in Neuropsychiatric Disorders through Monitoring of Arsenic and Its Derivatives as Well as Through Bioinformatics and Cheminformatics.Int J Mol Sci. 2019 Apr 12;20(8):1804. doi: 10.3390/ijms20081804. Int J Mol Sci. 2019. PMID: 31013686 Free PMC article. Review.
-
Computational ligand-based rational design: Role of conformational sampling and force fields in model development.Medchemcomm. 2011 May;2(5):356-370. doi: 10.1039/C1MD00044F. Medchemcomm. 2011. PMID: 21716805 Free PMC article.
References
-
- Bleicher K.H., Böhm H.-J., Müller K., Alanine A.I. Hit and lead generation: beyond high-throughput screening. Nat. Rev. Drug Discov. 2003;2:369–378. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

