Sampling the solution space in genome-scale metabolic networks reveals transcriptional regulation in key enzymes
- PMID: 20657658
- PMCID: PMC2904763
- DOI: 10.1371/journal.pcbi.1000859
Sampling the solution space in genome-scale metabolic networks reveals transcriptional regulation in key enzymes
Abstract
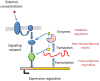
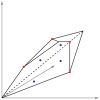
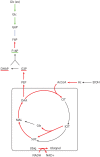
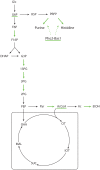
Genome-scale metabolic models are available for an increasing number of organisms and can be used to define the region of feasible metabolic flux distributions. In this work we use as constraints a small set of experimental metabolic fluxes, which reduces the region of feasible metabolic states. Once the region of feasible flux distributions has been defined, a set of possible flux distributions is obtained by random sampling and the averages and standard deviations for each of the metabolic fluxes in the genome-scale model are calculated. These values allow estimation of the significance of change for each reaction rate between different conditions and comparison of it with the significance of change in gene transcription for the corresponding enzymes. The comparison of flux change and gene expression allows identification of enzymes showing a significant correlation between flux change and expression change (transcriptional regulation) as well as reactions whose flux change is likely to be driven only by changes in the metabolite concentrations (metabolic regulation). The changes due to growth on four different carbon sources and as a consequence of five gene deletions were analyzed for Saccharomyces cerevisiae. The enzymes with transcriptional regulation showed enrichment in certain transcription factors. This has not been previously reported. The information provided by the presented method could guide the discovery of new metabolic engineering strategies or the identification of drug targets for treatment of metabolic diseases.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
-
- Yang C, Hua Q, Shimizu K. Integration of the information from gene expression and metabolic fluxes for the analysis of the regulatory mechanisms in Synechocystis. Appl Microbiol Biotechnol. 2002;58:813–822. - PubMed
-
- Fong SS, Nanchen A, Palsson BO, Sauer U. Latent pathway activation and increased pathway capacity enable Escherichia coli adaptation to loss of key metabolic enzymes. J Biol Chem. 2006;281:8024–8033. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

